- Record: found
- Abstract: found
- Article: found
Discovery of an Ebolavirus-Like Filovirus in Europe
Read this article at
Abstract
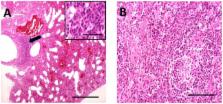
Filoviruses, amongst the most lethal of primate pathogens, have only been reported as natural infections in sub-Saharan Africa and the Philippines. Infections of bats with the ebolaviruses and marburgviruses do not appear to be associated with disease. Here we report identification in dead insectivorous bats of a genetically distinct filovirus, provisionally named Lloviu virus, after the site of detection, Cueva del Lloviu, in Spain.
Author Summary
A novel filovirus, provisionally named Lloviu virus (LLOV), was detected during the investigation of Miniopterus schreibersii die-offs in Cueva del Lloviu in southern Europe. LLOV is genetically distinct from other marburgviruses and ebolaviruses and is the first filovirus detected in Europe that was not imported from an endemic area in Africa. Filoviruses, amongst the most lethal of primate pathogens, have only been reported as natural infections in sub-Saharan Africa and the Philippines. Infections of bats with the ebolaviruses and marburgviruses do not appear to be associated with disease. Here we report identification of genetically distinct filovirus in dead insectivorous bats in caves in Spain.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Marburg Virus Infection Detected in a Common African Bat
- Record: found
- Abstract: found
- Article: not found
Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations.
- Record: found
- Abstract: found
- Article: not found