- Record: found
- Abstract: found
- Article: not found
Interfering with disease: a progress report on siRNA-based therapeutics
Read this article at
Abstract
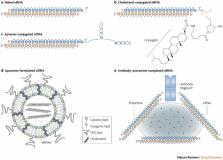
RNA interference (RNAi) has rapidly advanced since its initial discovery to form the basis of a new class of therapeutics. De Fougerolles and colleagues discuss the challenges in the development of RNAi-based therapeutics, focusing on lead identification/optimization and effective delivery, and review the latest clinical results.
Abstract
RNA interference (RNAi) quietly crept into biological research in the 1990s when unexpected gene-silencing phenomena in plants and flatworms first perplexed scientists. Following the demonstration of RNAi in mammalian cells in 2001, it was quickly realized that this highly specific mechanism of sequence-specific gene silencing might be harnessed to develop a new class of drugs that interfere with disease-causing or disease-promoting genes. Here we discuss the considerations that go into developing RNAi-based therapeutics starting from in vitro lead design and identification, to in vivo pre-clinical drug delivery and testing. We conclude by reviewing the latest clinical experience with RNAi therapeutics.
Related collections
Most cited references79
- Record: found
- Abstract: found
- Article: not found
Mechanisms of gene silencing by double-stranded RNA.
- Record: found
- Abstract: found
- Article: not found
Expression profiling reveals off-target gene regulation by RNAi.
- Record: found
- Abstract: found
- Article: not found
