- Record: found
- Abstract: found
- Article: found
Impact of swapping soils on the endophytic bacterial communities of pre-domesticated, ancient and modern maize
Read this article at
Abstract
Background
Endophytes are microbes that live within plants such as maize (corn, Zea mays L.) without causing disease. It is generally assumed that most endophytes originate from soil. If this is true, then as humans collected, domesticated, bred and migrated maize globally from its native Mexico, they moved the species away from its native population of endophyte donors. The migration of maize persists today, as breeders collect wild and exotic seed (as sources of diverse alleles) from sites of high genetic diversity in Mexico for breeding programs on distant soils. When transported to new lands, it is unclear whether maize permits only selective colonization of microbes from the Mexican soils on which it co-evolved, tolerates shifts in soil-derived endophytes, or prevents colonization of soil-based microbes in favour of seed-transmitted microbes. To test these hypotheses, non-sterilized seeds of three types of maize (pre-domesticated-Mexican, ancient-Mexican, modern-temperate) were planted side-by-side on indigenous Mexican soil, Canadian temperate soil or sterilized sand. The impact of these soil swaps on founder bacterial endophyte communities was tested using 16S-rDNA profiling, culturing and microbial trait phenotyping.
Results
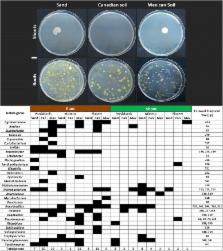
Multivariate analysis showed that bacterial 16S-rDNA TRFLP profiles from young, surface-sterilized maize plants were more similar when the same host genotype was grown on the different soils than when different maize genotypes were grown on the same soil. There appeared to be two reasons for this result. First, the largest fraction of bacterial 16S-signals from soil-grown plants was shared with parental seeds and/or plants grown on sterilized sand, suggesting significant inheritance of candidate endophytes. The in vitro activities of soil-derived candidate endophytes could be provided by bacteria that were isolated from sterile sand grown plants. Second, many non-inherited 16S-signals from sibling plants grown on geographically-distant soils were shared with one another, suggesting maize can select microbes with similar TRFLP peak sizes from diverse soils. Wild, pre-domesticated maize did not possess more unique 16S-signals when grown on its native Mexican soil than on Canadian soil, pointing against long-term co-evolutionary selection. The modern hybrid did not reject more soil-derived 16S-signals than did ancestral maize, pointing against such rejection as a mechanism that contributes to yield stability across environments. A minor fraction of 16S-signals was uniquely associated with any one soil.
Conclusion
Within the limits of TRFLP profiling, the candidate bacterial endophyte populations of pre-domesticated, ancient and modern maize are partially buffered against the effects of geographic migration --- from a Mexican soil associated with ancestral maize, to a Canadian soil associated with modern hybrid agriculture. These results have implications for understanding the effects of domestication, migration, ex situ seed conservation and modern breeding, on the microbiome of one of the world’s most important food crops.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
A single domestication for maize shown by multilocus microsatellite genotyping.
- Record: found
- Abstract: found
- Article: not found
The Diversity of Archaea and Bacteria in Association with the Roots of Zea mays L.
- Record: found
- Abstract: found
- Article: not found