- Record: found
- Abstract: found
- Article: found
Genome-Wide Analysis of Serine Hydroxymethyltransferase Genes in Triticeae Species Reveals That TaSHMT3A-1 Regulates Fusarium Head Blight Resistance in Wheat
Read this article at
Abstract
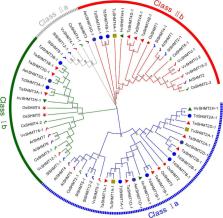
Serine hydroxymethyltransferase ( SHMT) plays a pivotal role in cellular one-carbon, photorespiration pathways and it influences the resistance to biotic and abiotic stresses. However, the function of SHMT proteins in wheat remains largely unexplored. In the present study, SHMT genes in five Triticeae species, Oryza sativa, and four dicotyledon species were identified based on whole genome information. The origin history of the target gene was traced by micro-collinearity analysis. Gene expression patterns of TaSHMTs in different tissues, various biotic stresses, exogenous hormones, and two biotic stresses were determined by Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). The function of the selected TaSHMT3A-1 was studied by barley stripe mosaic virus-induced gene silencing in common wheat Bainong207. A total of 64 SHMT members were identified and further classified into two main classes based on the structure of SHMT proteins. The gene structure and motif composition analyses revealed that SHMTs kept relatively conserved within the same subclasses. Interestingly, there was a gene, TdSHMT7B-1, on chromosome 7B of Triticum dicoccoides, but there was no SHMT gene on chromosome 7 of other analyzed Triticeae species; TdSHMT7B-1 had fewer exons and conserved motifs than the genes in the same subclass, suggesting that the gene of TdSHMT7B-1 has a notable evolutionary progress. The micro-collinearity relationship showed that no homologs of TaSHMT3A-1 and its two neighboring genes were found in the collinearity region of Triticum urartu, and there were 27 genes inserted into the collinearity region of T. urartu. Furthermore, qRT-PCR results showed that TaSHMT3A-1 was responsive to abiotic stresses (NaCl and cold), abscisic acid, methyl jasmonate, and hydrogen peroxide. Significantly, upon Fusarium graminearum infection, the expression of TaSHMT3A-1 was highly upregulated in resistant cultivar Sumai3. More importantly, silencing of TaSHMT3A-1 compromises Fusarium head blight resistance in common wheat Bainong207. Our new findings suggest that the TaSHMT3A-1 gene in wheat plays an important role in resistance to Fusarium head blight. This provides a valuable reference for further study on the function of this gene family.
Related collections
Most cited references63
- Record: found
- Abstract: found
- Article: not found
MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms.
- Record: found
- Abstract: found
- Article: not found
TBtools - an integrative toolkit developed for interactive analyses of big biological data

- Record: found
- Abstract: found
- Article: found