- Record: found
- Abstract: found
- Article: found
Screening and functional identification of lncRNAs in antler mesenchymal and cartilage tissues using high-throughput sequencing
Read this article at
Abstract
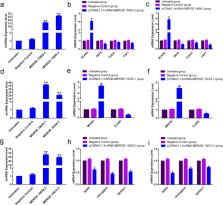
Long non-coding RNA (lncRNA) is a transcription product of the mammalian genome that regulates the development and growth in the body. The present study aimed to analyze the expression dynamics of lncRNA in sika antler mesenchymal and cartilage tissues by high-throughput sequencing. Bioinformatics was applied to predict differentially expressed lncRNAs and target genes and screen lncRNAs and mRNAs related to osteogenic differentiation, cell proliferation, and migration. Finally, the expression of the lncRNAs and target genes were analyzed by qRT-PCR. The results showed that compared to the cartilage tissue, the transcription levels of lncRNA and mRNA, 1212 lncRNAs and 518 mRNAs, in mesenchymal tissue were altered significantly. Thus, a complex interaction network was constructed, and the lncRNA-mRNA interaction network correlation related to osteogenic differentiation, cell proliferation, and migration was analyzed. Among these, the 26 lncRNAs and potential target genes were verified by qRT-PCR, and the results of qRT-PCR were consistent with high-throughput sequencing results. These data indicated that lncRNA promotes the differentiation of deer antler mesenchymal tissue into cartilage tissue by regulating the related osteogenic factors, cell proliferation, and migration-related genes and accelerating the process of deer antler regeneration and development.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
Evolution and functions of long noncoding RNAs.

- Record: found
- Abstract: found
- Article: found