- Record: found
- Abstract: found
- Article: found
TIME FOR COFFEE controls root meristem size by changes in auxin accumulation in Arabidopsis
Read this article at
Abstract
Reduced root meristem length in tic mutant is due to the repressed expression of PIN genes for decreased acropetal auxin transport, leading to low auxin accumulation. MYC2-mediated JA-signaling pathway may not be involved in this process
Abstract
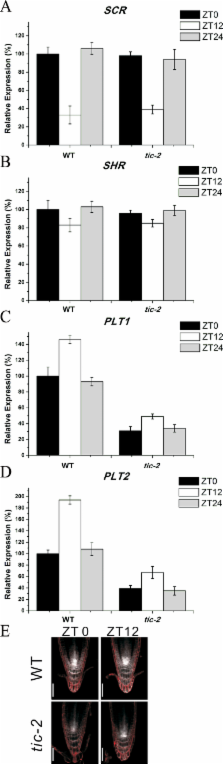
Roots play important roles in plant survival and productivity as they not only anchor the plants in the soil but are also the primary organ for the uptake of nutrients from the outside. The growth and development of roots depend on the specification and maintenance of the root meristem. Here, we report a previously unknown role of TIME FOR COFFEE ( TIC) in controlling root meristem size in Arabidopsis. The results showed that loss of function of TIC reduced root meristem length and cell number by decreasing the competence of meristematic cells to divide. This was due to the repressed expression of PIN genes for decreased acropetal auxin transport in tic-2, leading to low auxin accumulation in the roots responsible for reduced root meristem, which was verified by exogenous application of indole-3-acetic acid. Downregulated expression of PLETHORA1 ( PLT1) and PLT2, key transcription factors in mediating the patterning of the root stem cell niche, was also assayed in tic-2. Similar results were obtained with tic-2 and wild-type plants at either dawn or dusk. We also suggested that the MYC2-mediated jasmonic acid signalling pathway may not be involved in the regulation of TIC in controlling the root meristem. Taken together, these results suggest that TIC functions in an auxin– PLTs loop for maintenance of post-embryonic root meristem.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
Local, efflux-dependent auxin gradients as a common module for plant organ formation.
- Record: found
- Abstract: found
- Article: not found
The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots.
- Record: found
- Abstract: found
- Article: not found