- Record: found
- Abstract: found
- Article: found
Eugene – A Domain Specific Language for Specifying and Constraining Synthetic Biological Parts, Devices, and Systems
Read this article at
Abstract
Background
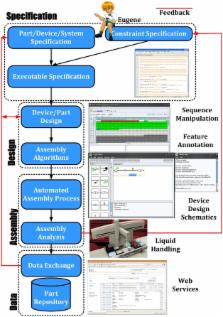
Synthetic biological systems are currently created by an ad-hoc, iterative process of specification, design, and assembly. These systems would greatly benefit from a more formalized and rigorous specification of the desired system components as well as constraints on their composition. Therefore, the creation of robust and efficient design flows and tools is imperative. We present a human readable language (Eugene) that allows for the specification of synthetic biological designs based on biological parts, as well as provides a very expressive constraint system to drive the automatic creation of composite Parts (Devices) from a collection of individual Parts.
Results
We illustrate Eugene's capabilities in three different areas: Device specification, design space exploration, and assembly and simulation integration. These results highlight Eugene's ability to create combinatorial design spaces and prune these spaces for simulation or physical assembly. Eugene creates functional designs quickly and cost-effectively.
Conclusions
Eugene is intended for forward engineering of DNA-based devices, and through its data types and execution semantics, reflects the desired abstraction hierarchy in synthetic biology. Eugene provides a powerful constraint system which can be used to drive the creation of new devices at runtime. It accomplishes all of this while being part of a larger tool chain which includes support for design, simulation, and physical device assembly.
Related collections
Most cited references38
- Record: found
- Abstract: found
- Article: not found
The second wave of synthetic biology: from modules to systems.
- Record: found
- Abstract: found
- Article: not found