- Record: found
- Abstract: found
- Article: found
A Time-Based and Intratumoral Proteomic Assessment of a Recurrent Glioblastoma Multiforme
Read this article at
Abstract
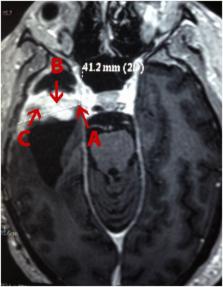
Tumors consist of cells in different stages of transformation with molecular and cellular heterogeneity. By far, heterogeneity is the hallmark of glioblastoma multiforme (GBM), the most malignant and aggressive type of glioma. Most proteomic studies aim in comparing tumors from different patients, but here we dive into exploring the intratumoral proteome diversity of a single GBM. For this, we profiled tumor fragments from the profound region of the same patient’s GBM but obtained from two surgeries a year’s time apart. Our analysis also included GBM‘s fragments from different anatomical regions. Our quantitative proteomic strategy employed 4-plex iTRAQ peptide labeling followed by a four-step strong cation chromatographic separation; each fraction was then analyzed by reversed-phase nano-chromatography coupled on-line with an Orbitrap-Velos mass spectrometer. Unsupervised clustering grouped the proteomic profiles into four major distinct groups and showed that most changes were related to the tumor’s anatomical region. Nevertheless, we report differentially abundant proteins from GBM’s fragments of the same region but obtained 1 year apart. We discuss several key proteins (e.g., S100A9) and enriched pathways linked with GBM such as the Ras pathway, RHO GTPases activate PKNs, and those related to apoptosis, to name a few. As far as we know, this is the only report that compares GBM fragments proteomic profiles from the same patient. Ultimately, our results fuel the forefront of scientific discussion on the importance in exploring the richness of subproteomes within a single tissue sample for a better understanding of the disease, as each tumor is unique.
Related collections
Most cited references51
- Record: found
- Abstract: found
- Article: not found
Evolutionary dynamics of carcinogenesis and why targeted therapy does not work.
- Record: found
- Abstract: found
- Article: found
Intratumor heterogeneity: seeing the wood for the trees.
- Record: found
- Abstract: found
- Article: not found