- Record: found
- Abstract: found
- Article: found
News in livestock research — use of Omics-technologies to study the microbiota in the gastrointestinal tract of farm animals
Read this article at
Abstract
Technical progress in the field of next-generation sequencing, mass spectrometry and bioinformatics facilitates the study of highly complex biological samples such as taxonomic and functional characterization of microbial communities that virtually colonize all present ecological niches. Compared to the structural information obtained by metagenomic analyses, metaproteomic approaches provide, in addition, functional data about the investigated microbiota. In general, integration of the main Omics-technologies (genomics, transcriptomics, proteomics and metabolomics) in live science promises highly detailed information about the specific research object and helps to understand molecular changes in response to internal and external environmental factors.
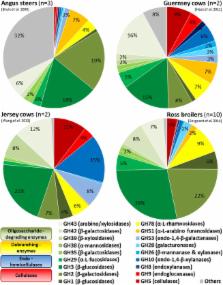
The microbial communities settled in the mammalian gastrointestinal tract are essential for the host metabolism and have a major impact on its physiology and health. The microbiotas of livestock like chicken, pig and ruminants are becoming a focus of interest for veterinaries, animal nutritionists and microbiologists. While pig is more often used as an animal model for human-related studies, the rumen microbiota harbors a diversity of enzymes converting complex carbohydrates into monomers which bears high potential for biotechnological applications.
This review will provide a general overview about the recent Omics-based research of the microbiota in livestock including its major findings. Differences concerning the results of pre- Omics-approaches in livestock as well as the perspectives of this relatively new Omics-platform will be highlighted.
Related collections
Most cited references77

- Record: found
- Abstract: found
- Article: found
Metagenomics - a guide from sampling to data analysis
- Record: found
- Abstract: found
- Article: not found
Diversity and succession of the intestinal bacterial community of the maturing broiler chicken.
- Record: found
- Abstract: found
- Article: not found