- Record: found
- Abstract: found
- Article: found
Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway
Read this article at
Abstract
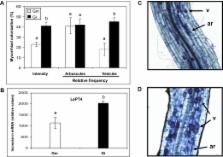
Arbuscular mycorrhizal (AM) symbioses are mutualistic associations between soil fungi and most vascular plants. The symbiosis significantly affects the host physiology in terms of nutrition and stress resistance. Despite the lack of host range specificity of the interaction, functional diversity between AM fungal species exists. The interaction is finely regulated according to plant and fungal characters, and plant hormones are believed to orchestrate the modifications in the host plant. Using tomato as a model, an integrative analysis of the host response to different mycorrhizal fungi was performed combining multiple hormone determination and transcriptional profiling. Analysis of ethylene-, abscisic acid-, salicylic acid-, and jasmonate-related compounds evidenced common and divergent responses of tomato roots to Glomus mosseae and Glomus intraradices, two fungi differing in their colonization abilities and impact on the host. Both hormonal and transcriptional analyses revealed, among others, regulation of the oxylipin pathway during the AM symbiosis and point to a key regulatory role for jasmonates. In addition, the results suggest that specific responses to particular fungi underlie the differential impact of individual AM fungi on plant physiology, and particularly on its ability to cope with biotic stresses.
Related collections
Most cited references64
- Record: found
- Abstract: found
- Article: not found
Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development.
- Record: found
- Abstract: found
- Article: not found
The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis.
- Record: found
- Abstract: not found
- Article: not found