- Record: found
- Abstract: found
- Article: found
Comprehensive detection and identification of bacterial DNA in the blood of patients with sepsis and healthy volunteers using next-generation sequencing method - the observation of DNAemia
Read this article at
Abstract
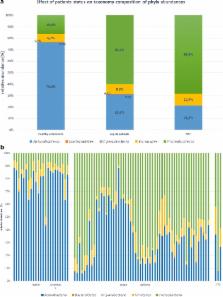
Blood is considered to be a sterile microenvironment, in which bacteria appear only periodically. Previously used methods allowed only for the detection of either viable bacteria with low sensitivity or selected species of bacteria. The Next-Generation Sequencing method (NGS) enables the identification of all bacteria in the sample with their taxonomic classification. We used NGS for the analysis of blood samples from healthy volunteers ( n = 23) and patients with sepsis ( n = 62) to check whether any bacterial DNA exists in the blood of healthy people and to identify bacterial taxonomic profile in the blood of septic patients. The presence of bacterial DNA was found both in septic and healthy subjects; however, bacterial diversity was significantly different ( P = 0.002) between the studied groups. Among healthy volunteers, a significant predominance of anaerobic bacteria (76.2 %), of which most were bacteria of the order Bifidobacteriales (73.0 %), was observed. In sepsis, the majority of detected taxa belonged to aerobic or microaerophilic microorganisms (75.1 %). The most striking difference was seen in the case of Actinobacteria phyla, the abundance of which was decreased in sepsis ( P < 0.001) and Proteobacteria phyla which was decreased in the healthy volunteers ( P < 0.001). Our research shows that bacterial DNA can be detected in the blood of healthy people and that its taxonomic composition is different from the one seen in septic patients. Detection of bacterial DNA in the blood of healthy people may suggest that bacteria continuously translocate into the blood, but not always cause sepsis; this observation can be called DNAemia.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection.
- Record: found
- Abstract: found
- Article: not found
Does blood of healthy subjects contain bacterial ribosomal DNA?
- Record: found
- Abstract: found
- Article: not found