- Record: found
- Abstract: found
- Article: found
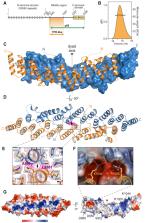
Structural basis for the dimerization of Gemin5 and its role in protein recruitment and translation control
research-article
Read this article at
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Abstract
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
TPR proteins: the versatile helix.
Luca D'Andrea, Lynne Regan (2003)
- Record: found
- Abstract: not found
- Article: not found
Dali: a network tool for protein structure comparison.
Liisa Holm, Chris Sander (1995)
- Record: found
- Abstract: found
- Article: not found
The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity
Deniz Simsek, Gerald C. Tiu, Ryan Flynn … (2017)