- Record: found
- Abstract: found
- Article: found
Highly Multiplexed Single-Cell In Situ RNA and DNA Analysis by Consecutive Hybridization
Read this article at
Abstract
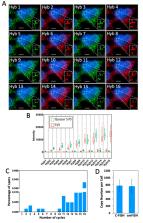
The ability to comprehensively profile nucleic acids in individual cells in their natural spatial contexts is essential to advance our understanding of biology and medicine. Here, we report a novel method for spatial transcriptomics and genomics analysis. In this method, every nucleic acid molecule is detected as a fluorescent spot at its natural cellular location throughout the cycles of consecutive fluorescence in situ hybridization (C-FISH). In each C-FISH cycle, fluorescent oligonucleotide probes hybridize to the probes applied in the previous cycle, and also introduce the binding sites for the next cycle probes. With reiterative cycles of hybridization, imaging and photobleaching, the identities of the varied nucleic acids are determined by their unique color sequences. To demonstrate the feasibility of this method, we show that transcripts or genomic loci in single cells can be unambiguously quantified with 2 fluorophores and 16 C-FISH cycles or with 3 fluorophores and 9 C-FISH cycles. Without any error correction, the error rates obtained using the raw data are close to zero. These results indicate that C-FISH potentially enables tens of thousands (2 16 = 65,536 or 3 9 = 19,683) of different transcripts or genomic loci to be precisely profiled in individual cells in situ.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
Proteomics. Tissue-based map of the human proteome.
- Record: found
- Abstract: found
- Article: not found
RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells.
- Record: found
- Abstract: found
- Article: not found