- Record: found
- Abstract: found
- Article: found
MicroRNA‐382 silencing induces a mitonuclear protein imbalance and activates the mitochondrial unfolded protein response in muscle cells
Read this article at
Abstract
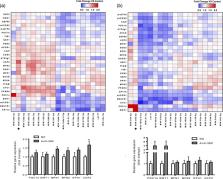
Proper mitochondrial function plays a central role in cellular metabolism. Various diseases as well as aging are associated with diminished mitochondrial function. Previously, we identified 19 miRNAs putatively involved in the regulation of mitochondrial metabolism in skeletal muscle, a highly metabolically active tissue. In the current study, these 19 miRNAs were individually silenced in C2C12 myotubes using antisense oligonucleotides, followed by measurement of the expression of 27 genes known to play a major role in regulating mitochondrial metabolism. Based on the outcomes, we then focused on miR‐382‐5p and identified pathways affected by its silencing using microarrays, investigated protein expression, and studied cellular respiration. Silencing of miRNA‐382‐5p significantly increased the expression of several genes involved in mitochondrial dynamics and biogenesis. Conventional microarray analysis in C2C12 myotubes silenced for miRNA‐382‐5p revealed a collective downregulation of mitochondrial ribosomal proteins and respiratory chain proteins. This effect was accompanied by an imbalance between mitochondrial proteins encoded by the nuclear and mitochondrial DNA (1.35‐fold, p < 0.01) and an induction of HSP60 protein (1.31‐fold, p < 0.05), indicating activation of the mitochondrial unfolded protein response (mtUPR). Furthermore, silencing of miR‐382‐5p reduced basal oxygen consumption rate by 14% ( p < 0.05) without affecting mitochondrial content, pointing towards a more efficient mitochondrial function as a result of improved mitochondrial quality control. Taken together, silencing of miR‐382‐5p induces a mitonuclear protein imbalance and activates the mtUPR in skeletal muscle, a phenomenon that was previously associated with improved longevity.
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
Adipose mitochondrial biogenesis is suppressed in db/db and high-fat diet-fed mice and improved by rosiglitazone.
- Record: found
- Abstract: found
- Article: not found