- Record: found
- Abstract: found
- Article: not found
Insight into the sequence specificity of a probe on an Affymetrix GeneChip by titration experiments using only one oligonucleotide
Read this article at
Abstract
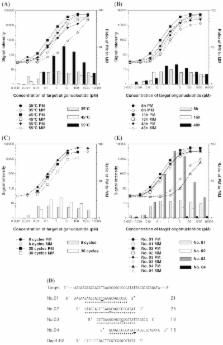
High-density oligonucleotide arrays are powerful tools for the analysis of genome-wide expression of genes and for genome-wide screens of genetic variation in living organisms. One of the critical problems in high-density oligonucleotide arrays is how to identify the actual amounts of a transcript due to noise and cross-hybridization involved in the observed signal intensities. Although mismatch (MM) probes are spotted on Affymetrix GeneChips to evaluate the noise and cross-hybridization embedded in perfect match (PM) probes, the behavior of probe-level signal intensities remains unclear. In the present study, we hybridized only one complement 25-mer oligonucleotide to characterize the behavior of duplex formation between target and probe in the complete absence of cross-hybridization. Titration experiments using only one oligonucleotide demonstrated that a substantial amount of intact target was hybridized not only to the PM but also the MM probe and that duplex formation between intact target and MM probe was efficiently reduced by increasing the stringency of hybridization conditions and shortening probe length. In addition, we discuss the correlation between potential for secondary structure of target oligonucleotide and hybridization intensity. These findings will be useful for the development of genome-wide analysis of gene expression and genetic variations by optimization of hybridization and probe conditions.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics.
- Record: found
- Abstract: found
- Article: not found
High density synthetic oligonucleotide arrays.
- Record: found
- Abstract: found
- Article: not found
