- Record: found
- Abstract: found
- Article: found
Computational models for predicting drug responses in cancer research
Read this article at
Abstract
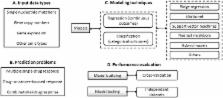
The computational prediction of drug responses based on the analysis of multiple types of genome-wide molecular data is vital for accomplishing the promise of precision medicine in oncology. This will benefit cancer patients by matching their tumor characteristics to the most effective therapy available. As larger and more diverse layers of patient-related data become available, further demands for new bioinformatics approaches and expertise will arise. This article reviews key strategies, resources and techniques for the prediction of drug sensitivity in cell lines and patient-derived samples. It discusses major advances and challenges associated with the different model development steps. This review highlights major trends in this area, and will assist researchers in the assessment of recent progress and in the selection of approaches to emerging applications in oncology.
Related collections
Most cited references49

- Record: found
- Abstract: found
- Article: found
pRRophetic: An R Package for Prediction of Clinical Chemotherapeutic Response from Tumor Gene Expression Levels
- Record: found
- Abstract: found
- Article: not found
The NCI60 human tumour cell line anticancer drug screen.

- Record: found
- Abstract: found
- Article: found