- Record: found
- Abstract: found
- Article: found
The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis
Read this article at
Abstract
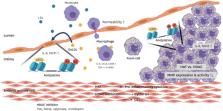
Atherosclerosis, which is the most common chronic disease of the coronary artery, constitutes a vascular pathology induced by inflammation and plaque accumulation within arterial vessel walls. Both DNA methylation and histone modifications are epigenetic changes relevant for atherosclerosis. Recent studies have shown that the DNA methylation and histone modification systems are closely interrelated and mechanically dependent on each other. Herein, we explore the functional linkage between these systems, with a particular emphasis on several recent findings suggesting that histone acetylation can help in targeting DNA methylation and that DNA methylation may control gene expression during atherosclerosis.
Related collections
Most cited references58
- Record: found
- Abstract: found
- Article: not found
TET enzymes, TDG and the dynamics of DNA demethylation.
- Record: found
- Abstract: found
- Article: not found
Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme.
- Record: found
- Abstract: found
- Article: not found