- Record: found
- Abstract: found
- Article: found
Mapping a Type-specific Epitope by Monoclonal Antibody against VP3 Protein of Duck Hepatitis A Type 1 Virus
Read this article at
Abstract
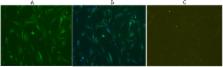
Duck hepatitis A subtype 1 virus (DHAV-1) infection causes high mortality in ducklings, resulting in significant losses to duck industries. VP3 is a structural protein of DHAV-1. However, B-cell epitopes on VP3 have not been investigated. To stimulate VP3 antibody response, eukaryotic expression plasmid pCI-neo-VP3 was constructed and used as DNA immunogen to prepare mAbs. Western blot showed that 25.5 kDa VP3 could be detected by mAbs in duck embryo fibroblast (DEF) cells transfected with pCI-neo-VP3. Immunofluorescence assay showed that mAbs could specifically bind to DEF cells infected with DHAV-1. DAPI staining indicated that VP3 localizes to the cytoplasm and nucleus of DHAV-1 infected DEF. With neutralizing mAb 3B7, minimal epitope PSNI was mapped. Sequence alignment indicated that 205PSNI 208 is highly conserved among DHAV-1, but different from those of DHAV-2 and DHAV-3. Epitope peptide reacted specifically with DHAV-1-positive duck sera by dot blotting, revealing PSNI is DHAV-1 type-specific epitope and the importance of these amino acids in antibody-epitope binding reactivity. These findings provided useful information for understanding the antigenicity of VP3 and might be valuable in the development of epitope-based vaccine or diagnostic kit for DHAV-1 infection and provide insights for understanding the pathogenesis of DHAV-1.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: not found
Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification.
- Record: found
- Abstract: found
- Article: not found
Nuclear targeting sequences--a consensus?
- Record: found
- Abstract: found
- Article: not found