- Record: found
- Abstract: found
- Article: found
The C–S–A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice
Read this article at
Abstract
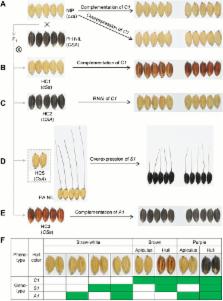
The C–S–A gene system determines rice hull pigmentation. A conserved color-producing model reveals the independent origin and evolution of the anthocyanin biosynthesis pathway in rice.
Abstract
Floral organs in rice ( Oryza sativa) can be purple, brown, or red in color due to the accumulation of flavonoids, but the molecular mechanism underlying specific organ pigmentation is not clear. Here, we propose a C–S–A gene model for rice hull pigmentation and characterize it through genetic, molecular, and metabolomic approaches. Furthermore, we conducted phylogenetic studies to reveal the evolution of rice color. In this gene system, C1 encodes a R2R3-MYB transcription factor and acts as a color-producing gene, and S1 encodes a bHLH protein that functions in a tissue-specific manner. C1 interacts with S1 and activates expression of A1, which encodes a dihydroflavonol reductase. As a consequence, the hull is purple where functional A1 participation leads to high accumulation of cyanidin 3- O-glucoside. Loss of function of A1 leads to a brown hull color due to accumulation of flavonoids such as hesperetin 5- O-glucoside, rutin, and delphinidin 3- O-rutinoside. This shows a different evolutionary pathway of rice color in japonica and indica, supporting independent origin of cultivars in each subspecies. Our findings provide a complete perspective on the gene regulation network of rice color formation and supply the theoretical basis for extended application of this beneficial trait.
Related collections
Most cited references33
- Record: found
- Abstract: found
- Article: not found
MEGA6: Molecular Evolutionary Genetics Analysis version 6.0.
- Record: found
- Abstract: found
- Article: not found
Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA.
- Record: found
- Abstract: found
- Article: not found