- Record: found
- Abstract: found
- Article: found
Cancer Stem Cells in Soft-Tissue Sarcomas
Read this article at
Abstract
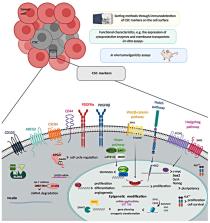
Soft tissue sarcomas (STS) are a rare group of mesenchymal solid tumors with heterogeneous genetic profiles and clinical features. Systemic chemotherapy is the backbone treatment for advanced STS; however, STS frequently acquire resistance to standard therapies, which highlights the need to improve treatments and identify novel therapeutic targets. Increases in the knowledge of the molecular pathways that drive sarcomas have brought to light different molecular alterations that cause tumor initiation and progression. These findings have triggered a breakthrough of targeted therapies that are being assessed in clinical trials. Cancer stem cells (CSCs) exhibit mesenchymal stem cell (MSC) features and represent a subpopulation of tumor cells that play an important role in tumor progression, chemotherapy resistance, recurrence and metastasis. In fact, CSCs phenotypes have been identified in sarcomas, allied to drug resistance and tumorigenesis. Herein, we will review the published evidence of CSCs in STS, discussing the molecular characteristic of CSCs, the commonly used isolation techniques and the new possibilities of targeting CSCs as a way to improve STS treatment and consequently patient outcome.
Related collections
Most cited references111
- Record: found
- Abstract: found
- Article: not found
Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation

- Record: found
- Abstract: found
- Article: found
Targeting mTOR for cancer therapy
- Record: found
- Abstract: found
- Article: found