- Record: found
- Abstract: not found
- Article: not found
3,3’-Diindolylmethane suppresses high-fat diet-induced obesity through inhibiting adipogenesis of pre-adipocytes by targeting USP2 activity
Hee Yang
1 ,
Sang Gwon Seo
1 ,
Seung Ho Shin
1 ,
Soyun Min
1 ,
Min Jeong Kang
1 ,
Ra Yoo
1 ,
Jeong Yeon Kwon
2 ,
Shuhua Yue
3 ,
Kee Hong Kim
2 ,
Ji-Xin Cheng
3
,
4 ,
Jong Rhan Kim
5 ,
Joon-Suk Park
6 ,
Jong Hun Kim
7 ,
Jung Han Yoon Park
7 ,
Hyong Joo Lee
1 ,
Ki Won Lee
1
,
7
,
8
July 18 2017
Read this article at
ScienceOpenPublisher
There is no author summary for this article yet. Authors can add summaries to their articles on ScienceOpen to make them more accessible to a non-specialist audience.
Related collections
Most cited references32
- Record: found
- Abstract: found
- Article: not found
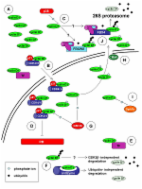
Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes.
- Record: found
- Abstract: found
- Article: found
The regulation of cyclin D1 degradation: roles in cancer development and the potential for therapeutic invention
John Alao (2007)
- Record: found
- Abstract: found
- Article: not found
Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway.
Eric J Diehl, C Sherr, F Zindy (1997)