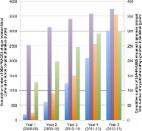
INTRODUCTION A growing body of literature has established the benefits of research experiences for undergraduate students in the sciences (Seymour et al., 2004; Lopatto, 2006, 2009; Laursen et al., 2010). Indeed, integration of research experiences into the academic-year curriculum, along with the use of other active-learning strategies, is a central theme in calls for undergraduate biology education reform. (See BIO2010: Transforming Undergraduate Education for Future Research Biologists [National Research Council (NRC), 2003] and Vision and Change in Undergraduate Biology Education: A Call to Action [American Association for the Advancement of Science, 2011].) Such experiences can be particularly beneficial for first-generation students, underrepresented minorities, and at-risk students, significantly improving retention rates in the sciences for these groups (Nagda et al., 1998; Hathaway et al., 2002; Lopatto, 2006, 2007; Locks and Gregerman, 2008; Goins et al., 2009). Perhaps as a consequence, the second recommendation in the recent President's Council of Advisors on Science and Technology (PCAST) report, Engage to Excel (2012, pp. 16, 25, and 38), is to “advocate and provide support for replacing standard laboratory courses with discovery-based research courses.” Utilizing results from a diverse group of institutions, the growth in the Genomics Education Partnership (GEP) provides us with the opportunity to examine key features of undergraduate instruction that can contribute to student gains from a research experience embedded in an academic-year class (Shaffer et al., 2010). Importantly, we find that these results are independent of institution characteristics. Rather, the key variable is the length of time spent on the project. Genomics studies as a focus for undergraduate research provide many opportunities, and are especially useful for teaching institutions limited by minimal research infrastructure and budgetary support. First, there is a huge amount of raw genomics data, most archived and publicly available (National Center for Biotechnology Information [NCBI], www.ncbi.nlm.nih.gov). Most of this sequence data has been analyzed only by individual prediction programs at the time of posting, providing ample opportunities for undergraduates to improve this analysis and carry out their own investigations mining information from genomes. “Turning data into knowledge” (Brenner, 2002) is in fact the major bottleneck in genomics today, and a large-scale undergraduate project in sequence improvement and analysis results in new understandings and improved data sets that can benefit the research community as a whole (Boomer et al., 2002; Elwess and Latourelle, 2004; Drew and Triplett, 2008). Further, with DNA sequencing continuously becoming less expensive, we can anticipate many more institutions initiating genome-sequencing or RNA-sequencing projects in the not-too-distant future, allowing students to explore local ecosystems and characterize genomes of local organisms (e.g., see Oleksyk et al., 2012). In addition, the “tools of the trade” in genomics and bioinformatics are generally publicly available (e.g., at the NCBI) and lend themselves well to peer instruction. Today's students are often very adept at using computers, and students who are familiar with the computer-based tools of bioinformatics from previous experience prove to be effective coaches for newcomers, serving as peer instructors or teaching assistants (TAs). Peers can promote the development of a dynamic undergraduate research community in which students can serve as scientific mentors, with attendant benefits (Harrison et al., 2010; Dunbar et al., 2012). A faculty member with the requisite background in the fundamentals of genetics, molecular biology, and/or evolutionary biology can coach students to think critically about their observations and to question, probe, and analyze the data to address relevant scientific questions. Together, faculty and peer instructors can create the foundation for a lively genomics research team that is actively contributing to the scientific body of knowledge. Success in this project can provide students with the increased confidence that is a hallmark of research experiences for undergraduates. In the National Survey of Student Engagement (Bennett et al., 2007), students deem research experience to be a high-impact practice. To this end, several national programs have been established during the past few years that take advantage of genomics to engage undergraduates in research (Hanauer et al., 2006; Hatfull et al., 2006; Campbell et al., 2007; Hingamp et al., 2008; Shaffer et al., 2010; Ditty et al., 2010; Banta et al., 2012; Singer et al., 2013), in keeping with the recommendations of PCAST (2012), Vision and Change (AAAS, 2011), and BIO2010 (NRC, 2003). Despite these reported successes and positive reports from long-range studies (Bauer and Bennett, 2003; Brodl, 2005), we have encountered some skepticism as to whether basing curriculum on a research project is an approach that is an efficient use of time and/or is broadly applicable to a diverse population of students, or whether it is best reserved for high-achieving students working in the summer, the current tradition. The GEP provides an opportunity to evaluate the efficacy of a research project specifically in genomics in both attaining student learning objectives and achieving informed student attitudes regarding research experiences, drawing from a diverse pool of students. The GEP is designed to engage undergraduates in a joint research project in genomics while introducing them to bioinformatics tools and resources with the goal of increasing their understanding of eukaryotic genes and genomes, as well as immersing them in the practice of science. This project, initiated in 2006, now has ∼100 affiliated schools and has engaged more than 1000 students at ∼60 different colleges and universities during the past year alone. Training materials to familiarize students with bioinformatics tools and relevant strategies have been developed at Washington University and at GEP member schools, with collaborative assessment and revision. Undergraduate students participating in the program can improve the quality of genomic sequence and annotate genes and other features, elucidating meaning from DNA sequence. Research questions in genomics are addressed using the results of these efforts, leading to student presentations both locally and nationally, and ultimately research publications. Since 2006, students involved in the program have worked with the Genome Institute at Washington University in St. Louis (WUSTL) to improve more than 7 million bases of draft genomic sequence from several species of Drosophila. Using a suite of bioinformatics tools selected and/or developed in collaboration between the Department of Biology and Department of Computer Science and Engineering at Washington University (often with additional input generated on their campus), students have produced hundreds of gene models using evidence-based manual annotation. Improved sequences are submitted to GenBank (2013) and used in studies exploring genome evolution (e.g., see Leung et al., 2010; other manuscripts are currently in preparation). Since our initial assessment of students enrolled in 2008–2009 (Shaffer et al., 2010), the GEP has doubled in size and attracted a diverse group of schools in terms of size, educational mission, and public versus private support. Faculty members have collaboratively developed a variety of ways to use the GEP approach in their teaching, including short (∼10 h) modules in a genetics course, longer modules within molecular biology laboratory courses, stand-alone genomics lab courses, and independent research studies. This diversity of schools and approaches has allowed us to look for critical variables for student success, measured both in terms of responses to an online SURE-style survey (SURE is the Survey of Undergraduate Research Experiences [Lopatto, 2004, 2007]), which we will refer to as the “learning survey,” as well as an online knowledge-based quiz. In addition, to determine long-term impacts on students’ subsequent actions and careers, we have surveyed occupations and attitudes of students one or more years after the completion of a GEP-affiliated course. We find that institutional characteristics have little correlation with student success, indicating that diverse students in diverse settings benefit from curriculum-based research experiences of this type. We see a similar impact on student attitudes from the GEP course compared with a traditional independent summer research experience as measured by the learning survey, but find that the impact correlates with the amount of time the instructor was able to devote to the project. The impact of time spent is seen not only with students at the end of their GEP course experience, but also in their reports on their experiences in subsequent years. The data make a strong argument for allotting more instructional time to research-based work in the undergraduate curriculum. MATERIALS AND METHODS The Student Research Project The GEP is a collaborative effort between the Department of Biology, the Department of Computer Science and Engineering, and the Genome Institute, all located at WUSTL, and a growing number of colleges and universities. (See http://gep.wustl.edu/community/current_members for a list of currently affiliated schools, and Figure 6 later in this article for characteristics of the schools participating in this assessment.) The project is organized around a central database housed and maintained by WUSTL on a pair of SUSE Enterprise Linux servers that host a variety of services utilized by the GEP community, including curriculum modules, access to research projects and bioinformatics tools, and assessment and communications tools (see http://gep.wustl.edu and Shaffer et al., 2010). Figure 1. The GEP sequence improvement and annotation workflow. All projects are completed at least twice independently, and the results are reconciled before final assembly and analysis. While the quality of the genome sequence assembly available is sufficient to proceed directly to annotation for some species, in other cases it is necessary to improve the genome assembly (“finishing”) by generating additional sequencing data and manually resolving misassemblies. Figure 2. The GEP UCSC Genome Browser. Students are challenged to analyze and evaluate the available evidence assembled on the genome browser to create optimal gene models and explore other genomic features. A sample of the available tracks is shown here. Often the available evidence is contradictory; e.g., see the discrepancy between some of the gene predictions and the organization of genes suggested by homology with D. melanogaster (BLASTX alignments). Figure 3. GEP students show gains in their understanding of genes and genomes. The bars depict the mean scores on a 20-point quiz on genes and genomes attained by GEP students who participated in annotation and by a comparison group over 2010–2011 and 2011–2012. The GEP pretest data include 1026 observations; the GEP posttest data include 748 observations. The increase in scores was evaluated in two ways: first, with an independent-groups t test (t = 26.1, df = 1836, p 36 h) is required to obtain the full benefits of a research experience. Figure 8. Comparison of student responses on the 20 learning gain items (mean and SEM) from the SURE survey. The data are classified by instructor reports of the number of hours devoted to the annotation project. These were divided into four quartiles as shown in Figure 7; the responses from the Q1 (1–10 h) and Q4 (>36 h) students are shown here. The Q1 group includes 86–112 observations; the Q4 group includes 149–175 observations. (Respondents tallied here were those who answered the specific question on the SURE survey.) Error bars represent 2 SEM. Figure 9. Comparison of student responses on seven additional learning benefit items categorized by the reported number of hours of instruction and lab work on the annotation project (Q1: 1–10 h; Q4: > 36 h). The Q1 group includes 105–107 observations; the Q4 group includes 172–174 observations. (Respondents tallied here were those who answered the specific question.) The error bars represent two SEM. Alumni Attitudes Also Show Increased Value with More Time Invested To determine the effect of this curriculum-based research experience on student career trajectories, we obtained demographic and attitudinal data on students who had formerly taken a GEP course or served as a TA in a GEP course. Data were collected during the summer and fall of 2012 by an online survey that included demographic and attitudinal questions. The earliest cohort surveyed took GEP classes in 2005, the last cohort in Fall 2011. We had 473 valid responses (29% of the students contacted); however, because all questions were voluntary, the total number of responses to any one question rarely totaled 473. The students reported that they came from 41 different institutions. The diversity of institutions represented is similar to that found in the student survey (Figure 6; see data in Figure S2). Of the respondents, 200 described themselves as male, 268 as female; the pool was 59% Caucasian, 20% Asian, 17% underrepresented minorities (African American, African, Hispanic), and 5% mixed plus other. All others declined to respond. To investigate the current occupations of the alumni respondents, we gave them a series of 16 career categories and allowed students to select one or more as appropriate. Figure 10 shows the total number of alumni that selected each category. The three largest areas of occupation (pooling some categories) reported by the alumni were being a postsecondary student in science (pursuing an MA, PhD, or other professional degree: 30%), medical school student (pursuing an MD or MD/PhD: 22%), and employment in science (19%). Only 9% indicated they were no longer in science (pooled PhD nonscience, other professional nonscience, and employed nonscience). These results are very encouraging; however, as in any survey of this type, the findings may be impacted by response bias; that is, those students continuing in science may have been more inclined to respond to the request to participate in the survey. Figure 10. The current occupations of GEP alumni as self-reported on the alumni survey. To examine the attitudes these alumni had toward science in general and their experience with the GEP in particular, we asked them to complete a survey much the same as our original postcourse learning survey. Alumni were asked to reflect on and evaluate their experiences and learning gains, given that some time had passed since they had taken the GEP course. Figure 11 shows the average response to these questions, with 1 being “strongly disagree” and 5 being “strongly agree,” that their GEP experience fulfilled each particular goal. In addition to the overall average for these topics, we also cross-correlated the results with the extent of the GEP experience to which each student was exposed. Students were asked to pick one of four responses as describing the type of GEP course that they took. These were: 1) a course devoted primarily to the GEP project, 2) a course spending half or more of the time on the GEP project, 3) a course that spent a quarter of the time on the GEP project, and 4) a course in which just one to three labs were devoted to the GEP project. Figure 11 shows the average responses for each of these groups (numerical data in Table S2). In general, there was a clear and consistent increase in attitudinal gains as students spent more course time working on their GEP project. The average of all items for each group was 1) 4.02, 2) 3.62, 3) 3.30, and 4) 3.03. Figure 11. Alumni student attitudes separated by extent of GEP experience. Purple: “Took up just 1–3 labs”; green: “Course spent quarter”; red: “Course spent half or more”; blue: “Course devoted primarily to the GEP project.” Scale is 1–5, with 1 being “strongly disagree” and 5 being “strongly agree.” See Table S2 for numerical values. Items common to both the alumni and current student surveys allow us to directly compare responses from current 2012–2013 GEP students with those of the alumni. Figure 12 shows these comparison data. In most cases, the alumni had a very positive response to their GEP experience, showing an average response to each item that was ∼0.25–0.4 units higher than the average from the current GEP 2012 cohort. One item, “Genomics is awesome,” showed a difference larger than the typical range, with a difference of 0.63. These results suggest that the students value the experience more as they find that what they learned in their annotation project has applicability to their further pursuits. Figure 12. Comparison of alumni and current student responses to six attitudinal survey questions. Scale is 1–5, with 1 being “strongly disagree” and 5 being “strongly agree.” Again, one might be concerned about response bias. In particular, we asked whether the students who had spent more time working on the GEP research project were disproportionately represented in the pool of students who responded to the alumni survey. We found that 30% of the alumni respondents fell into the group having a brief GEP experience (one to three lab sessions, the equivalent of Q1 above), while 26% of the current students fall into Q1 (<10 h work with GEP). Thus, the two pools are roughly similar in distribution across the course types through which the GEP project is offered. Alumni students were also invited to comment on their GEP experience. In particular, students were invited to respond to the question “Please comment on your GEP experience: What was good about it, and what changes would have made it better?” Most of the student comments were favorable (∼90%). The most common comments from alumni stated that they enjoyed the course, they enjoyed the independence, they appreciated having a real research project, they learned a lot, and the experience made them feel like researchers. Other comments emphasized that the experience was relevant to their future plans, was interesting, and inspired teamwork. Several students commented on their initial confusion and subsequent development of understanding. (See Table 3 for a numerical analysis of the 320 comments, as well as sample comments for each category.) The results of this survey support the conclusion that we can provide a robust learning experience in genomics through a research project. They further reinforce the notion that an in-depth research experience with a considerable time investment produces the most tangible and long-term gains. Table 3. Frequency analysis of alumni responses to the question “Please comment on your GEP experience: What was good about it, and what changes would have made it better?” Comment Frequency Examplesa Enjoyment (literally saying “I enjoyed the course” or a thought much like it). 42 I enjoyed getting to experience the ownership of a portion of a project (my fosmid) while still being able to have the opportunity to work in a group setting and learn with my peers. I enjoyed participating in the GEP program because it allowed myself and a partner to really take a hands on approach to genomic education. The project was ours, had our name on it and we really felt like we contributed to the science. The process was long and at times difficult but overall, I enjoyed being part of it. I really enjoyed the feeling of participating in a meaning [sic] research project. In addition, I think it encouraged active participation in the class and encouraged partners to work diligently together to achieve a common goal. Research-like (comments about how either the topic or the work made the student feel like a researcher as opposed to a passive student). 10 Working on near-independent projects in the small-group setting really set the course apart from other college experiences; it is certainly the closest a course ever came to emulating an actual research project. I liked that we were doing real, primary research, working directly with original sequence data. It was nice to actually be doing science “first-hand.” Important or significant (similar to faculty comments that it was useful to be engaged in significant research). 6 I liked that what I did actually counted! It was very rewarding to take a class in which your work truly has an impact on the scientific community in comparison to most other classes where the learning is essentially to achieve personal means of learning the information and performing well on the examinations. I thought it was great to contribute to something practical and useful. Confusion to clarity (comments that the early part of the course was confusing or frustrating but then learning led to clarity and understanding). 6 From what I recall, it was initially confusing as I got familiar with the websites and software required to analyze fosmids. Once I got going, though, it was like solving a puzzle, and I was satisfied with my work when the project was over. I can't think of anything in particular I would like to change. Frustration can easily occur when looking for a your fosmid. However, struggling through this research is also the best part because it is the best way to learn and understand your research. Constantly it was a trial and error for figuring out which key to use or tab, but once I knew how to use it, it was very easy but for the first couple of times it was confusing. Relevant (was relevant to science or to grad school or career). 8 I think genomics is becoming a more relevant field and every student with an interest in bioinformatics, cell/molecular biology, or genetics should be exposed to the GEP course. I really enjoyed learning how to use all of the online bioinformatics resources. They have been extremely useful in my graduate studies. I think a stronger emphasis on annotation should be made due to the advances in sequencing technology [that] will probably make finishing obsolete in the future. The GEP experience was very helpful in introducing me to the field of bioinformatics and its associated tools. I consistently make use of the skills I gained while taking the course now, during the completion of my PhD program. It also helped to improve communicating scientific data, as I presented my results during the class and will also be a coauthor in an upcoming scientific article. Realistic (the research was on a “real” or “authentic” problem as opposed to a scripted lab). 16 I liked that we were doing real, primary research, working directly with original sequence data. Gave a chance to get students involved in real-world work in the field of molecular biology and bioinformatics. Interesting and a great way to problem solve. Having a hand in generating real primary research as an undergraduate gave me so much more independence in my view of science. I did feel like I owned my project and was contributing to the greater field of knowledge. Independence (working and thinking independently). 22 It really involved a lot of independent thought and problem solving ability. I enjoyed the challenge and I enjoyed the fact that I was contributing to the knowledge base. I truly enjoyed working independently on our GEP project. It was the only course I had in my undergraduate career that had novel research findings and did not require work with a partner. It was somewhat difficult to keep a “notebook” during the course, though I believe that this is a reflection of how parts of research are becoming more computational and are not as conducive to a traditional daily log of research. Working independently (separate from the professor) on a project where the answers were not previously known was an invaluable lesson about the scientific process. Learned a lot. 14 I learned more about genomics in this class than I did in any other class I took in school. I really did learn an immense amount of information while taking the course. Before taking this course, I had no idea how to annotate genes of any specimen and had never heard of websites such as pubmed, NCBI, flybase, etc. This GEP course really exposed me to a whole new topic within what we had learned in high school. I really enjoyed being able to understand the complex, yet stimulating GEP information in this course. I learned a lot about genomics, bioinformatics tools and about research. I think that it was a great experience. Interesting. 5 The experience that I received increased my interest in bioinformatics. It was interesting to learn how to use the interactive tools and fun to discover new genes. It made me feel as if I was a part of something important. At first, I was somewhat confused as to what it was that we (my partner and I) had to do, but with time it became more and more interesting and fun. Teamwork is important. 4 Teamwork was good and the feeling of contributing to science. Working as a team with my partner was fun. I learned a lot about sequencing. Great experience of working together as a team and the results were perfect. Negative (comments about time and clarity of instruction). 36 We should have spent more time doing the project. It was frustrating to spend all that time learning how to complete the project and then only doing one fosmid. I feel there needed to be more time spent introducing and really explaining the subject matter. I felt thrown into it and it only tangentially related to my course material. The entire process should have been explained more. I was not sure what I was doing. aVerbatim sample quotes are shown for each category. DISCUSSION The importance of providing research experiences for science, technology, engineering, and mathematics undergraduates is well documented (see references cited in the Introduction). However, providing such experiences has been challenging due to the substantial resource and personnel requirements for individual mentored research. Classroom-based undergraduate research experiences can provide an alternative, but despite this, many outstanding pilot projects (see Introduction) have not been widely adopted, perhaps because of concerns about the different needs and cultures of institutions of various sizes, missions, and budget, or of students with diverse levels of preparation. The expansion of the GEP to ∼100 affiliated schools in the past 3 yr has allowed for the collection and analysis of student outcomes to determine whether specific factors contribute to the knowledge gains (quizzes) and self-reported learning gains in understanding research (survey) observed among GEP students. Institutional characteristics such as public or private funding, student body size, the presence or absence of advanced degree programs in biology, or the prevalence of particular subgroups of students such as first-generation college students or minority students had no significant effect on the overall knowledge and learning survey scores. Interestingly, responses by both current students and GEP alumni indicate that gains are enhanced by increased time invested in the research project in the classroom. These findings demonstrate the utility of a broad-based, centrally organized genomics research project in providing successful classroom research experiences for undergraduates across diverse institutions. They further suggest that it is worthwhile to devote a significant amount of class time to achieve the full benefits of a research experience for a large number of students. The outcomes of student participation in such a program compare favorably with extracurricular undergraduate research experiences in terms of positive influence on students over time. This outcome may in part be the result of features of the project structure that mirror both the scientific process and utilization of the cognitive skills outlined in Bloom's taxonomy; student responses indicate strong gains, particularly in learning about the nature of science and the research process, along with attainment of competence and confidence in research skills, particularly in analyzing conflicting data and achieving a defendable resolution. The data demonstrate a significant correlation between the amount of time devoted in a course to GEP materials and the perception of the experience by undergraduates as actual research, a result seen among both current students and GEP alumni. Importantly, students who were exposed to as little as 10 total semester hours of GEP work showed significant learning gains, as did students exposed to an average of 45.5 instructional hours (Q4) in a semester. However, the distinction between smaller and larger faculty investments of class time in the research project is revealed when assessing student confidence data; a significant investment of classroom time in the project is necessary to provide an experience that attains the full benefits of a research experience. This time-dependent variation in our learning versus confidence outcomes reflects the classic question of “content versus experience” that is at the heart of many pedagogical debates in science education: Is the student educational experience compromised by sacrificing emphasis on content for a more inquiry-based and experiential learning? We would argue, based on our above findings, that the use of a research project as a significant component of a biology course has significantly bolstered student confidence and competence, as evidenced by GEP alumni student comments and by them continuing their careers in the sciences. An increase in the science-literate workforce is one of the most tangible goals as we respond to calls for increased undergraduate research opportunities. A notable outcome of implementation of the GEP curriculum as compared with a SURE experience is the same or larger reported student learning gains in understanding the practice of science. The differences in scores between GEP and SURE students may be influenced by the fact that students in a research-based classroom environment are in a large community of peer learners. Undergraduate students involved in a summer research experience at a university are often working with a postdoc or graduate student and few direct peers. The parallel research experiences provided as part of a research-based course allow for peers to assist and support one another in deeper and more meaningful ways than they might otherwise have the opportunity to do. The simultaneous act of mentoring and being mentored contributes to the opportunities that students have to gain practice and confidence in their abilities as scientists. Additionally, not all undergraduate students involved in a SURE program have a substantial stake in the research project. Indeed, student comments on their GEP experiences often stress the importance of being responsible for their own projects, while participating as a team member. Finally, more than 90% of the implementations of the GEP curriculum required students to report their results in some manner to a broader scientific community, either to their class, to others at their institution, or to those assembled for regional or national scientific meetings. This opportunity for students to participate in the scientific community, as scientists do, contributes much toward shifting students from their accustomed roles as observers of the scientific process to active and competent participants in the scientific process. Clearly, we have not yet fully explored questions of scale. Those GEP-affiliated courses that strive for a full research experience are typically laboratory courses that enroll between five and 32 students, or independent study/research courses that enroll two to five students working together. Some GEP faculty members have used the project to provide an introduction to bioinformatics and gene annotation within a midlevel course in genetics enrolling 100–200 students. The instances to date devote only ∼10 h to the GEP project, so presumably have a lower impact, as documented above. Anecdotal observations suggest that one trained individual for every six to seven novices is required to avoid the inevitable frustrations that come with learning any new computational system, and implementation on a large scale therefore requires a cadre of instructors. While the participation of one or more senior scientists who can provide perspective and context for the research itself is an important part of the cohort of mentors, much of the mentoring required is procedural and is often best supplied by peer instructors (undergraduate TAs) who can easily communicate the ins and outs of computational infrastructure. How large a class could grow and still show significant student gains is unknown. Given appropriate staffing, and a willingness to devote class/lab time to the effort, it should be possible to expand the research experience to hundreds of students. How best to organize a large cohort to preserve the sense of a learning community and ensure that all students gain a sense of increased efficacy in doing science, has not yet been explored. What sorts of projects are well suited for a research-based laboratory course that can be implemented at a range of different institutional types? Ideally, one would like to see a range of projects of this type available, to provide a good match with the different research interests of faculty members. Our thoughts have been shaped by discussions with other faculty members who are experimenting with this style of teaching, particularly with HHMI professors Utpal Banerjee (see Chen et al., 2005; Call et al., 2007; Evans et al., 2009), Graham Hatfull (see Hanauer et al., 2006; Jacob-Sera et al., 2012), and Scott Strobel (see Bascom-Slack et al., 2012). The following concepts, highlighted by the work of Hatfull et al. (2006, see Table 3 therein), fit many undergraduate programs but apply to bioinformatics particularly well. The most important characteristic to consider in designing a research-based course is the development of a parallel project, one that allows the instructor to teach the students a common set of research tools and approaches while providing individual projects for the students. Ideally, the findings of the group as a whole provide added value to the individual results (e.g., the data will be more meaningful when brought together). For instance, given a starting DNA sequence assembly, a region of interest can be readily divided up among a class into smaller, overlapping projects, and the results subsequently used to reassemble the whole. The combined work of GEP students has allowed us to improve and provide careful annotation of megabase regions of selected eukaryotic genomes, something that could not be efficiently accomplished in any other way. Next, technical simplicity must be considered to maintain safety, provide compatibility with scheduling constraints, and ensure a high probability of success. A genomics research project has low start-up costs, requiring only computers with Internet access. There are no safety issues, and access to the project can be provided 24/7. There are no scheduling demands as such—no overnight incubations, no generation times to wait for, and so on. Many experiments, being electronic, can be repeated quickly in real time, so failure does not result in any significant costs or time penalties, and there is a high probability that one will learn something of value in the process. Third, a genomics research project can be designed at many levels, depending on the level of the students. For example, college freshmen generally do very well in annotating a prokaryotic genome or the genome of a phage (Hatfull et al., 2006; Caruso et al., 2009; Harrison et al., 2011) while upperclassmen are ready to annotate and analyze a eukaryotic genome with its additional complexities, including multi-exon genes (Leung et al., 2010; Shaffer et al., 2010). Once students are engaged in the project, they find that there are always more questions to ask. Students taking computer science as well as biology can generate their own programs to address questions of interest. A research course can also be designed to teach students skills in accessing various databases and using bioinformatics tools appropriately, with homework or quizzes to test those skills. We find that it is important to utilize various checkpoints (e.g., requiring a report on the first attempt to annotate a gene) to provide feedback to students and develop their confidence. Also, if one introduces students to the basic tools of the trade (e.g., EMBOSS, 2013; NCBI, 2013; UCSC Genome Browser, 2013; etc.), the skills they develop can of course be applied subsequently to analysis of other genes and genomes. Importantly, all research has as its goal publishable original findings, and this aspect gives students an added sense of responsibility, knowing that other scientists will build on their work; the prospect of a coauthorship provides a tangible reward for their efforts. Even if time does not permit commitment to the research goal, introducing students to the research tools and challenging them to analyze raw data are of value. Finally, the effort involved in completing one's own project, while part of a group receiving similar training and struggling with similar challenges, appears to generate student ownership of the projects and creates a dynamic partnership overall. Similar projects to the Drosophila work described here could be constructed around any number of model organisms to provide an accurate annotation of genomic regions of special interest and/or to address specific questions concerning the organization and evolution of genes and genomes (for additional examples, see Kerfeld and Simons, 2007; Ditty et al., 2010; Goff et al., 2011; Banta et al., 2012; Singer et al., 2013). A consortium clearly has cost/benefit advantages, and we believe that more national projects of this type should be supported by agencies interested in improving undergraduate science education in the United States in partnership with those interested in the daunting task of improving the utility of large data sets such as genomic sequences. Both the need for and the value of bona fide research experiences for undergraduates are well established, and the success of the GEP experience confirms that such experiences can be provided for students economically in terms of both time and money. The continued success of such efforts generally encourages us to believe that such in-class research will become an increasingly common part of the undergraduate science experience. Supplementary Material Supplemental Material