- Record: found
- Abstract: found
- Article: found
Physicochemical mechanisms of protein regulation by phosphorylation
Read this article at
Abstract
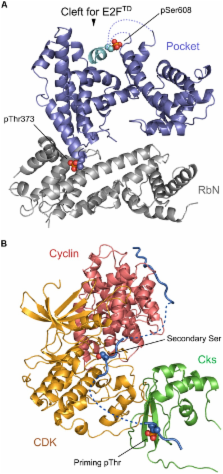
Phosphorylation offers a dynamic way to regulate protein activity and subcellular localization, which is achieved through its reversibility and fast kinetics. Adding or removing a dianionic phosphate group somewhere on a protein often changes the protein’s structural properties, its stability and dynamics. Moreover, the majority of signaling pathways involve an extensive set of protein–protein interactions, and phosphorylation can be used to regulate and modulate protein–protein binding. Losses of phosphorylation sites, as a result of disease mutations, might disrupt protein binding and deregulate signal transduction. In this paper we focus on the effects of phosphorylation on protein stability, dynamics, and binding. We describe several physico-chemical mechanisms of protein regulation through phosphorylation and pay particular attention to phosphorylation in protein complexes and phosphorylation in the context of disorder–order and order–disorder transitions. Finally we assess the role of multiple phosphorylation sites in a protein molecule, their possible cooperativity and function.
Related collections
Most cited references75
- Record: found
- Abstract: found
- Article: not found
The protein kinase complement of the human genome.
- Record: found
- Abstract: found
- Article: not found
Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm.
- Record: found
- Abstract: found
- Article: not found