- Record: found
- Abstract: found
- Article: found
Diversity and evolution of the small multidrug resistance protein family
Read this article at
Abstract
Background
Members of the small multidrug resistance (SMR) protein family are integral membrane proteins characterized by four α-helical transmembrane strands that confer resistance to a broad range of antiseptics and lipophilic quaternary ammonium compounds (QAC) in bacteria. Due to their short length and broad substrate profile, SMR proteins are suggested to be the progenitors for larger α-helical transporters such as the major facilitator superfamily (MFS) and drug/metabolite transporter (DMT) superfamily. To explore their evolutionary association with larger multidrug transporters, an extensive bioinformatics analysis of SMR sequences (> 300 Bacteria taxa) was performed to expand upon previous evolutionary studies of the SMR protein family and its origins.
Results
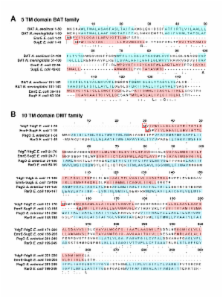
A thorough annotation of unidentified/putative SMR sequences was performed placing sequences into each of the three SMR protein subclass designations, namely small multidrug proteins (SMP), suppressor of groEL mutations (SUG), and paired small multidrug resistance (PSMR) using protein alignments and phylogenetic analysis. Examination of SMR subclass distribution within Bacteria and Archaea taxa identified specific Bacterial classes that uniquely encode for particular SMR subclass members. The extent of selective pressure acting upon each SMR subclass was determined by calculating the rate of synonymous to non-synonymous nucleotide substitutions using Syn-SCAN analysis. SUG and SMP subclasses are maintained under moderate selection pressure in comparison to integron and plasmid encoded SMR homologues. Conversely, PSMR sequences are maintained under lower levels of selection pressure, where one of the two PSMR pairs diverges in sequence more rapidly than the other. SMR genomic loci surveys identified potential SMR efflux substrates based on its gene association to putative operons that encode for genes regulating amino acid biogenesis and QAC-like metabolites. SMR subclass protein transmembrane domain alignments to Bacterial/Archaeal transporters (BAT), DMT, and MFS sequences supports SMR participation in multidrug transport evolution by identifying common TM domains.
Related collections
Most cited references64
- Record: found
- Abstract: found
- Article: not found
Multiple sequence alignment with the Clustal series of programs.
- Record: found
- Abstract: found
- Article: not found
Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria.
- Record: found
- Abstract: not found
- Article: not found