- Record: found
- Abstract: found
- Article: not found
A Genomic Approach to Identify Regulatory Nodes in the Transcriptional Network of Systemic Acquired Resistance in Plants
Read this article at
Abstract
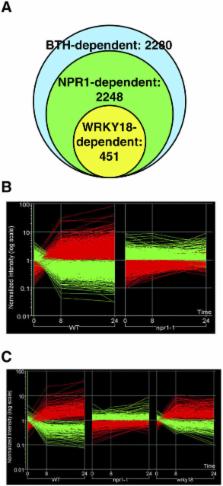
Many biological processes are controlled by intricate networks of transcriptional regulators. With the development of microarray technology, transcriptional changes can be examined at the whole-genome level. However, such analysis often lacks information on the hierarchical relationship between components of a given system. Systemic acquired resistance (SAR) is an inducible plant defense response involving a cascade of transcriptional events induced by salicylic acid through the transcription cofactor NPR1. To identify additional regulatory nodes in the SAR network, we performed microarray analysis on Arabidopsis plants expressing the NPR1-GR (glucocorticoid receptor) fusion protein. Since nuclear translocation of NPR1-GR requires dexamethasone, we were able to control NPR1-dependent transcription and identify direct transcriptional targets of NPR1. We show that NPR1 directly upregulates the expression of eight WRKY transcription factor genes. This large family of 74 transcription factors has been implicated in various defense responses, but no specific WRKY factor has been placed in the SAR network. Identification of NPR1-regulated WRKY factors allowed us to perform in-depth genetic analysis on a small number of WRKY factors and test well-defined phenotypes of single and double mutants associated with NPR1. Among these WRKY factors we found both positive and negative regulators of SAR. This genomics-directed approach unambiguously positioned five WRKY factors in the complex transcriptional regulatory network of SAR. Our work not only discovered new transcription regulatory components in the signaling network of SAR but also demonstrated that functional studies of large gene families have to take into consideration sequence similarity as well as the expression patterns of the candidates.
Synopsis
Many biological processes are controlled by intricate regulatory networks of gene expression. Identifying the regulatory nodes in these networks and understanding the hierarchical relationship between them are vital to our understanding of biological systems. However, this task is frequently hampered by the intrinsic complexity of these processes. Here, the authors used a controlled transcriptional profiling strategy to a plant immune response called systemic acquired resistance to study the transcriptional events one at a time. Systemic acquired resistance is activated through the induction of thousands of genes by the transcriptional regulator protein NPR1. The authors found that downstream of NPR1 are several regulatory nodes comprised of members from a large family of transcriptional factors. Disrupting these regulatory nodes compromised various functions assigned to NPR1, providing the information needed to construct a gene regulation network.
Related collections
Most cited references17
- Record: found
- Abstract: found
- Article: not found
DAVID: Database for Annotation, Visualization, and Integrated Discovery.
- Record: found
- Abstract: found
- Article: not found
Isochorismate synthase is required to synthesize salicylic acid for plant defence.
- Record: found
- Abstract: found
- Article: not found
