- Record: found
- Abstract: found
- Article: found
Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil

Read this article at
Significance
The diversity and ecology of RNA viruses is severely understudied in complex environments. Here we investigate the diversity and community patterns of soil RNA viruses by analyzing assembled metatranscriptomes. We tracked RNA viral and host communities for 22 d in 2 soil environments central to carbon cycling, the rhizosphere and detritosphere. This work is an important step toward understanding the factors that drive RNA viral communities. The main hosts in our system may be Fungi and Proteobacteria; this is in contrast to the ocean, where diatoms and other single cellular eukaryotes are primary hosts for RNA viruses. These results greatly expand the known diversity of viruses and contribute to understanding microbial interactions in soil with major implications for carbon cycling.
Abstract
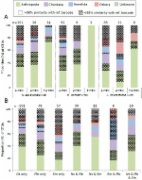
Viruses impact nearly all organisms on Earth, with ripples of influence in agriculture, health, and biogeochemical processes. However, very little is known about RNA viruses in an environmental context, and even less is known about their diversity and ecology in soil, 1 of the most complex microbial systems. Here, we assembled 48 individual metatranscriptomes from 4 habitats within a planted soil sampled over a 22-d time series: Rhizosphere alone, detritosphere alone, rhizosphere with added root detritus, and unamended soil (4 time points and 3 biological replicates). We resolved the RNA viral community, uncovering a high diversity of viral sequences. We also investigated possible host organisms by analyzing metatranscriptome marker genes. Based on viral phylogeny, much of the diversity was Narnaviridae that may parasitize fungi or Leviviridae, which may infect Proteobacteria. Both host and viral communities appear to be highly dynamic, and rapidly diverged depending on experimental conditions. The viral and host communities were structured based on the presence of root litter. Clear temporal dynamics by Leviviridae and their hosts indicated that viruses were replicating. With this time-resolved analysis, we show that RNA viruses are diverse, abundant, and active in soil. When viral infection causes host cell death, it may mobilize cell carbon in a process that may represent an overlooked component of soil carbon cycling.
Related collections
Most cited references74
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found