- Record: found
- Abstract: found
- Article: found
Integrated Transcriptome and Pathway Analyses Revealed Multiple Activated Pathways in Breast Cancer
Read this article at
Abstract
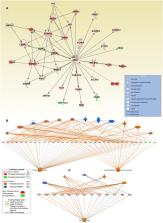
Breast cancer (BC) is the leading cause of cancer-related death in women. Therefore, a better understanding of BC biology and signaling pathways might lead to the development of novel biomarkers and targeted therapies. Although a number of transcriptomic studies have been performed on breast cancer patients from various geographic regions, there are almost no such comprehensive studies performed on breast cancer from patients in the gulf region. This study aimed to provide a better understanding of the altered molecular networks in BC from the gulf region. Herein, we compared the transcriptome of BC to adjacent normal tissue from six BC patients and identified 1,108 upregulated and 518 downregulated transcripts. A selected number of genes from the RNA-Seq analysis were subsequently validated using qRT-PCR. Differentially expressed (2.0-fold change, adj. p < 0.05) transcripts were subjected to ingenuity pathway analysis, which revealed a myriad of affected signaling pathways and functional categories. Activation of ERBB2, FOXM1, ESR1, and IGFBP2 mechanistic networks was most prominent in BC tissue. Additionally, BC tissue exhibited marked enrichment in genes promoting cellular proliferation, migration, survival, and DNA replication and repair. The presence of genes indicative of immune cell infiltration and activation was also observed in BC tissue. We observed high concordance [43.5% (upregulated) and 62.1% (downregulated)] between differentially expressed genes in our study group and those reported for the TCGA BC cohort. Our data provide novel insight on BC biology and suggest common altered molecular networks in BC in this geographic region. Our data suggest future development of therapeutic interventions targeting those common signaling pathways.
Related collections
Most cited references31
- Record: found
- Abstract: found
- Article: not found
A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer.
- Record: found
- Abstract: not found
- Article: not found
Chemokines and Chemokine Receptors: New Targets for Cancer Immunotherapy

- Record: found
- Abstract: found
- Article: found