- Record: found
- Abstract: found
- Article: not found
Reducing IRS-1 Activation Cause Mutation of Tyrosine Kinase Domain hINSR Gene on Type-2 Diabetes Mellitus Patients
Read this article at
Abstract
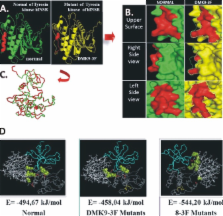
The purpose of this study is to examine the effect of mutation on tyrosine kinase hINSR gene of DM Type 2 patients reduce the IRS- 1 activation by in silico analysis. Blood DNA of DM Type 2 patients from Saeful Anwar Hospital Malang were amplified and sequenced by specific primers of tyrosine kinase domain of hINSR gene. These gene sequences were converted to protein sequence by BLAST and the IRS-1 protein sequence is retrieved from NCBI database. Both of the protein sequence was aligned by using Bio edit version 5.0.6. The model of three dimension protein was predicted by SWISS MODEL webserver, and visualized the structure alteration by using Pymol 0.99rc6 and Hex 5.0, and then superimpose of the hINSR and IRS-1 interaction were examined by docking using Hex 5.0. The results showed that one substitution and one deletion of 8-3F patient exon-22 hINSR gene tyrosine kinase domain cause loss of four helixes and three coils structures on tyrosine kinase hINSR protein. Six-deletions and six-substitutions on same gene domain of DMK9 patient changed the two helixes became coil structure. The binding energy of hINSR tyrosine kinase with IRS-1 of normal is E= -494.67 kJ/mol, DMK9 patient is E= -458.4 kJ/mol, and 8-3F patient is E=-544.20 kJ/mol. The DMK9 patient prognosis has better physiological condition than 8-3F patient. Interaction between 8-3F of hINSR tyrosine kinase domain mutation and PTB domain IRS-1 is more spontaneous than DMK9, but both of them were reduced on IRS-1 activation respectively.
Related collections
Most cited references14

- Record: found
- Abstract: found
- Article: found
The SWISS-MODEL Repository and associated resources

- Record: found
- Abstract: found
- Article: found
HexServer: an FFT-based protein docking server powered by graphics processors
- Record: found
- Abstract: found
- Article: not found
