- Record: found
- Abstract: found
- Article: not found
Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study
Read this article at
Summary
Background
In countries with declining numbers of confirmed cases of COVID-19, lockdown measures are gradually being lifted. However, even if most physical distancing measures are continued, other public health measures will be needed to control the epidemic. Contact tracing via conventional methods or mobile app technology is central to control strategies during de-escalation of physical distancing. We aimed to identify key factors for a contact tracing strategy to be successful.
Methods
We evaluated the impact of timeliness and completeness in various steps of a contact tracing strategy using a stochastic mathematical model with explicit time delays between time of infection and symptom onset, and between symptom onset, diagnosis by testing, and isolation (testing delay). The model also includes tracing of close contacts (eg, household members) and casual contacts, followed by testing regardless of symptoms and isolation if testing positive, with different tracing delays and coverages. We computed effective reproduction numbers of a contact tracing strategy ( R CTS) for a population with physical distancing measures and various scenarios for isolation of index cases and tracing and quarantine of their contacts.
Findings
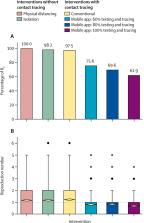
For the most optimistic scenario (testing and tracing delays of 0 days and tracing coverage of 100%), and assuming that around 40% of transmissions occur before symptom onset, the model predicts that the estimated effective reproduction number of 1·2 (with physical distancing only) will be reduced to 0·8 (95% CI 0·7–0·9) by adding contact tracing. The model also shows that a similar reduction can be achieved when testing and tracing coverage is reduced to 80% ( R CTS 0·8, 95% CI 0·7–1·0). A testing delay of more than 1 day requires the tracing delay to be at most 1 day or tracing coverage to be at least 80% to keep R CTS below 1. With a testing delay of 3 days or longer, even the most efficient strategy cannot reach R CTS values below 1. The effect of minimising tracing delay (eg, with app-based technology) declines with decreasing coverage of app use, but app-based tracing alone remains more effective than conventional tracing alone even with 20% coverage, reducing the reproduction number by 17·6% compared with 2·5%. The proportion of onward transmissions per index case that can be prevented depends on testing and tracing delays, and given a 0-day tracing delay, ranges from up to 79·9% with a 0-day testing delay to 41·8% with a 3-day testing delay and 4·9% with a 7-day testing delay.
Interpretation
In our model, minimising testing delay had the largest impact on reducing onward transmissions. Optimising testing and tracing coverage and minimising tracing delays, for instance with app-based technology, further enhanced contact tracing effectiveness, with the potential to prevent up to 80% of all transmissions. Access to testing should therefore be optimised, and mobile app technology might reduce delays in the contact tracing process and optimise contact tracing coverage.
Related collections
Most cited references26
- Record: found
- Abstract: found
- Article: found
Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China
- Record: found
- Abstract: found
- Article: not found
Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study
- Record: found
- Abstract: found
- Article: not found
