- Record: found
- Abstract: found
- Article: found
Engineering Yarrowia lipolytica for Campesterol Overproduction

Read this article at
Abstract
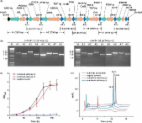
Campesterol is an important precursor for many sterol drugs, e. g. progesterone and hydrocortisone. In order to produce campesterol in Yarrowia lipolytica, C-22 desaturase encoding gene ERG5 was disrupted and the heterologous 7-dehydrocholesterol reductase (DHCR7) encoding gene was constitutively expressed. The codon-optimized DHCR7 from Rallus norvegicus, Oryza saliva and Xenapus laevis were explored and the strain with the gene DHCR7 from X. laevis achieved the highest titer of campesterol due to D409 in substrate binding sites. In presence of glucose as the carbon source, higher biomass conversion yield and product yield were achieved in shake flask compared to that using glycerol and sunflower seed oil. Nevertheless, better cell growth rate was observed in medium with sunflower seed oil as the sole carbon source. Through high cell density fed-batch fermentation under carbon source restriction strategy, a titer of 453±24.7 mg/L campesterol was achieved with sunflower seed oil as the carbon source, which is the highest reported microbial titer known. Our study has greatly enhanced campesterol accumulation in Y. lipolytica, providing new insight into producing complex and desired molecules in microbes.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: found
DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways
- Record: found
- Abstract: found
- Article: not found
Yarrowia lipolytica as a model for bio-oil production.
- Record: found
- Abstract: found
- Article: not found