- Record: found
- Abstract: found
- Article: found
Specific TATAA and bZIP requirements suggest that HTLV-I Tax has transcriptional activity subsequent to the assembly of an initiation complex
Read this article at
Abstract
Background
Human T-cell leukemia virus type I (HTLV-I) Tax protein is a transcriptional regulator of viral and cellular genes. In this study we have examined in detail the determinants for Tax-mediated transcriptional activation.
Results
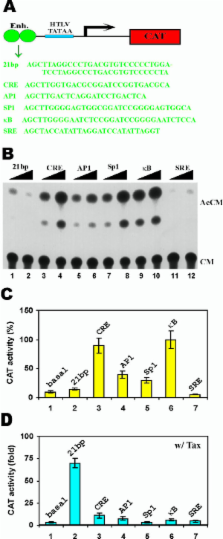
Whereas previously the LTR enhancer elements were thought to be the sole Tax-targets, herein, we find that the core HTLV-I TATAA motif also provides specific responsiveness not seen with either the SV40 or the E1b TATAA boxes. When enhancer elements which can mediate Tax-responsiveness were compared, the authentic HTLV-I 21-bp repeats were found to be the most effective. Related bZIP factors such as CREB, ATF4, c-Jun and LZIP are often thought to recognize the 21-bp repeats equivalently. However, amongst bZIP factors, we found that CREB, by far, is preferred by Tax for activation. When LTR transcription was reconstituted by substituting either κB or serum response elements in place of the 21-bp repeats, Tax activated these surrogate motifs using surfaces which are different from that utilized for CREB interaction. Finally, we employed artificial recruitment of TATA-binding protein to the HTLV-I promoter in "bypass" experiments to show for the first time that Tax has transcriptional activity subsequent to the assembly of an initiation complex at the promoter.
Related collections
Most cited references59
- Record: found
- Abstract: not found
- Article: not found