- Record: found
- Abstract: found
- Article: not found
Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences
Read this article at
Highlights
-
•
Recombination is very relevant in generating genetic variability in viral populations.
-
•
Viral recombination has important consequences for different areas of research. These include molecular biology, virology and evolutionary biology. It also impacts the daily practice of clinicians and public health officials.
-
•
Here we review three important aspects of viral recombination: (i) molecular mechanisms of model DNA- and RNA-viruses, (ii) methods for detection, characterization and quantification in viral populations, (iii) its impact on the evolutionary analysis of viral populations.
Abstract
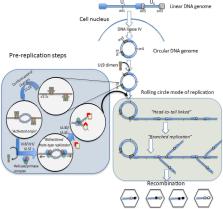
Recombination is a pervasive process generating diversity in most viruses. It joins variants that arise independently within the same molecule, creating new opportunities for viruses to overcome selective pressures and to adapt to new environments and hosts. Consequently, the analysis of viral recombination attracts the interest of clinicians, epidemiologists, molecular biologists and evolutionary biologists. In this review we present an overview of three major areas related to viral recombination: (i) the molecular mechanisms that underlie recombination in model viruses, including DNA-viruses (Herpesvirus) and RNA-viruses (Human Influenza Virus and Human Immunodeficiency Virus), (ii) the analytical procedures to detect recombination in viral sequences and to determine the recombination breakpoints, along with the conceptual and methodological tools currently used and a brief overview of the impact of new sequencing technologies on the detection of recombination, and (iii) the major areas in the evolutionary analysis of viral populations on which recombination has an impact. These include the evaluation of selective pressures acting on viral populations, the application of evolutionary reconstructions in the characterization of centralized genes for vaccine design, and the evaluation of linkage disequilibrium and population structure.
Related collections
Most cited references122
- Record: found
- Abstract: found
- Article: not found
The fine-scale structure of recombination rate variation in the human genome.
- Record: found
- Abstract: found
- Article: not found
Analyzing the mosaic structure of genes.
- Record: found
- Abstract: not found
- Article: not found
