- Record: found
- Abstract: found
- Article: not found
The Activity of HDAC8 Depends on Local and Distal Sequences of Its Peptide Substrates†

Read this article at
Abstract
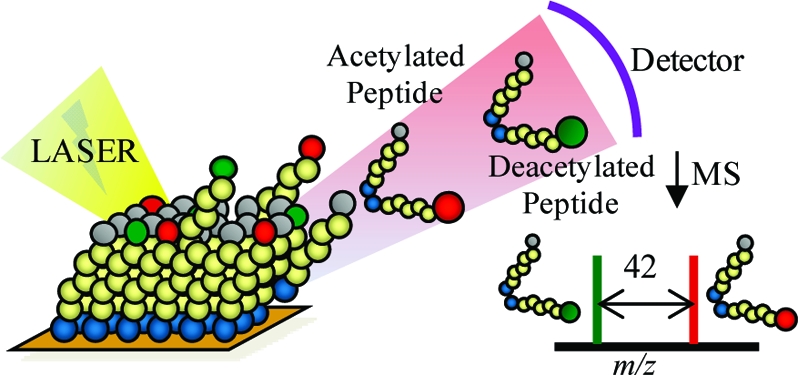
This paper introduces a flexible assay for characterizing the activities of the histone deacetylase enzymes. The approach combines mass spectrometry with self-assembled monolayers that present acetylated peptides and enables a label-free and one-step assay of this biochemical activity. The assay was used to characterize the activity of HDAC8 toward peptides taken from the N-terminal tail of the H4 histone and reveals that a distal region of the peptide substrate interacts with the deacetylase at an exosite and contributes to the activity of the substrate. Specifically, a peptide corresponding to residues 8−19 of H4 and having lysine 12 acetylated is an active substrate, but removal of the KRHR (residues 16−19) sequence abolishes activity. Mutation of glycine 11 to arginine in the peptide lacking the KRHR sequence restores activity, demonstrating that both local and distal sequences act synergistically to regulate the activity of the HDAC. Assays with peptides bearing multiply acetylated residues, but in which each acetyl group is isotopically labeled, permit studies of the processive deacetylation of peptides. Peptide substrates having an extended sequence that includes K20 were used to demonstrate that methylation of this residue directly affects HDAC8 activity at K12. This work provides a mechanistic basis for the regulation of HDAC activities by distal sequences and may contribute to studies aimed at evaluating the role of the histone code in regulating gene expression.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Structure and ligand of a histone acetyltransferase bromodomain.
- Record: found
- Abstract: not found
- Article: not found
ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS.
- Record: found
- Abstract: not found
- Article: not found
