- Record: found
- Abstract: found
- Article: not found
Orthotopic Patient-Derived Xenografts of Pediatric Solid Tumors
Abstract
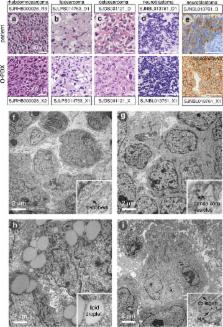
Pediatric solid tumors arise from endodermal, ectodermal, or mesodermal lineages 1 . Although the overall survival of children with solid tumors is 75%, that of children with recurrent disease is below 30% 2 . To capture the complexity and diversity of pediatric solid tumors and establish new models of recurrent disease, we developed a protocol to produce orthotopic patient-derived xenografts (O-PDXs) at diagnosis, recurrence, and autopsy. Tumor specimens were received from 168 patients, and 67 O-PDXs were established for 12 types of cancer. The origins of the O-PDX tumors were reflected in their gene-expression profiles and epigenomes. Genomic profiling of the tumors, including detailed clonal analysis, was performed to determine whether the clonal population in the xenograft recapitulated the patient’s tumor. We identified several drug vulnerabilities and showed that the combination of a WEE1 inhibitor (AZD1775), irinotecan, and vincristine can lead to complete response in multiple rhabdomyosarcoma O-PDX tumors in vivo.
Related collections
Most cited references15
- Record: found
- Abstract: not found
- Article: not found
Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing
- Record: found
- Abstract: found
- Article: not found
Organoid cultures derived from patients with advanced prostate cancer.
- Record: found
- Abstract: not found
- Article: not found
