- Record: found
- Abstract: found
- Article: found
Developmental Expression of Claudins in the Mammary Gland
Read this article at
Abstract
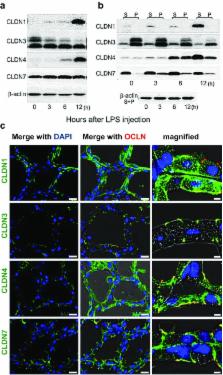
Claudins are a large family of membrane proteins whose classic function is to regulate the permeability of tight junctions in epithelia. They are tetraspanins, with four alpha-helices crossing the membrane, two extracellular loops, a short cytoplasmic N-terminus and a longer and more variable C-terminus. The extracellular ends of the helices are known to undergo side-to-side ( cis) interactions that allow the formation of claudin polymers in the plane of the membrane. The extracellular loops also engage in head-to-head ( trans) interactions thought to mediate the formation of tight junctions. However, claudins are also present in intracellular structures, thought to be vesicles, with less well-characterized functions. Here, we briefly review our current understanding of claudin structure and function followed by an examination of changes in claudin mRNA and protein expression and localization through mammary gland development. Claudins-1, 3, 4, 7, and 8 are the five most prominent members of the claudin family in the mouse mammary gland, with varied abundance and intracellular localization during the different stages of post-pubertal development. Claudin-1 is clearly localized to tight junctions in mammary ducts in non-pregnant non-lactating animals. Cytoplasmic puncta that stain for claudin-7 are present throughout development. During pregnancy claudin-3 is localized both to the tight junction and basolaterally while claudin-4 is found only in sparse puncta. In the lactating mouse both claudin-3 and claudin-8 are localized at the tight junction where they may be important in forming the paracellular barrier. At involution and under challenge by lipopolysaccharide claudins −1, −3, and −4 are significantly upregulated. Claudin-3 is still colocalized with tight junction molecules but is also distributed through the cytoplasm as is claudin-4. These largely descriptive data provide the essential framework for future mechanistic studies of the function and regulation of mammary epithelial cell claudins.
Related collections
Most cited references75
- Record: found
- Abstract: found
- Article: not found
Claudin-1 and -2: Novel Integral Membrane Proteins Localizing at Tight Junctions with No Sequence Similarity to Occludin
- Record: found
- Abstract: found
- Article: not found
Claudin-based tight junctions are crucial for the mammalian epidermal barrier
- Record: found
- Abstract: found
- Article: not found