- Record: found
- Abstract: found
- Article: found
Bacteriocins of lactic acid bacteria: extending the family
Read this article at
Abstract
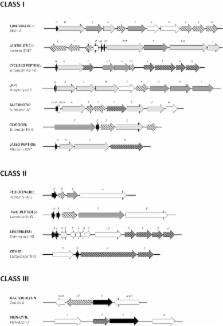
Lactic acid bacteria (LAB) constitute a heterogeneous group of microorganisms that produce lactic acid as the major product during the fermentation process. LAB are Gram-positive bacteria with great biotechnological potential in the food industry. They can produce bacteriocins, which are proteinaceous antimicrobial molecules with a diverse genetic origin, posttranslationally modified or not, that can help the producer organism to outcompete other bacterial species. In this review, we focus on the various types of bacteriocins that can be found in LAB and the organization and regulation of the gene clusters responsible for their production and biosynthesis, and consider the food applications of the prototype bacteriocins from LAB. Furthermore, we propose a revised classification of bacteriocins that can accommodate the increasing number of classes reported over the last years.
Related collections
Most cited references113
- Record: found
- Abstract: found
- Article: not found
Bacteriocins: developing innate immunity for food.
- Record: found
- Abstract: found
- Article: not found
antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers
- Record: found
- Abstract: found
- Article: not found