- Record: found
- Abstract: found
- Article: found
Mangrove-Associated Fungal Communities Are Differentiated by Geographic Location and Host Structure
Read this article at
Abstract
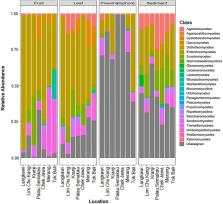
Marine fungi on the whole remain understudied, especially in the highly diverse Southeast Asian region. We investigated the fungal communities associated with the mangrove tree Avicennia alba throughout Singapore and Peninsular Malaysia. At each sampling location, we examined ten individual trees, collecting leaves, fruits, pneumatophores, and an adjacent sediment sample from each plant. Amplicon sequencing of the fungal internal transcribed spacer 1 and subsequent analyses reveal significant differences in fungal communities collected from different locations and host structures. Mantel tests and multiple regression on distance matrices show a significant pattern of distance decay with samples collected close to one another having more similar fungal communities than those farther away. Submergence appears to drive part of the variation as host structures that are never submerged (leaves and fruits) have more similar fungal communities relative to those that are covered by water during high tide (pneumatophores and sediment). We suggest that fungi of terrestrial origins dominate structures that are not inundated by tidal regimes, while marine fungi dominate mangrove parts and sediments that are submerged by the incoming tide. Given the critical functions fungi play in all plants, and the important role they can have in determining the success of restoration schemes, we advocate that fungal community composition should be a key consideration in any mangrove restoration or rehabilitation project.
Related collections
Most cited references54
- Record: found
- Abstract: not found
- Book Chapter: not found
AMPLIFICATION AND DIRECT SEQUENCING OF FUNGAL RIBOSOMAL RNA GENES FOR PHYLOGENETICS
- Record: found
- Abstract: not found
- Article: not found
VARIATION IN PLANT RESPONSE TO NATIVE AND EXOTIC ARBUSCULAR MYCORRHIZAL FUNGI

- Record: found
- Abstract: found
- Article: found