- Record: found
- Abstract: found
- Article: found
The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance
Read this article at
Abstract
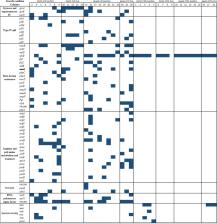
Ciprofloxacin (CIP) is used to treat Pseudomonas aeruginosa biofilm infections. We showed that the pathways of CIP-resistance development during exposure of biofilms and planktonic P. aeruginosa populations to subinhibitory levels of CIP depend on the mode of growth. In the present study, we analyzed CIP-resistant isolates obtained from previous evolution experiments, and we report a variety of evolved phenotypic and genotypic changes that occurred in parallel with the evolution of CIP-resistance. Cross-resistance to beta-lactam antibiotics was associated with mutations in genes involved in cell-wall recycling ( ftsZ, murG); and could also be explained by mutations in the TCA cycle ( sdhA) genes and in genes involved in arginine catabolism. We found that CIP-exposed isolates that lacked mutations in quorum-sensing genes and acquired mutations in type IV pili genes maintained swarming motility and lost twitching motility, respectively. Evolved CIP-resistant isolates showed high fitness cost in planktonic competition experiments, yet persisted in the biofilm under control conditions, compared with ancestor isolates and had an advantage when exposed to CIP. Their persistence in biofilm competition experiments in spite of their fitness cost in planktonic growth could be explained by their prolonged lag-phase. Interestingly, the set of mutated genes that we identified in these in vitro-evolved CIP-resistant colonies, overlap with a large number of patho-adaptive genes previously reported in P. aeruginosa isolates from cystic fibrosis (CF) patients. This suggests that the antibiotic stress is contributing to the bacterial evolution in vivo, and that adaptive laboratory evolution can be used to predict the in vivo evolutionary trajectories.
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
Microbiological effects of sublethal levels of antibiotics.

- Record: found
- Abstract: found
- Article: found
Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections
- Record: found
- Abstract: found
- Article: not found