- Record: found
- Abstract: found
- Article: found
Crystal Structure of Entamoeba histolytica Cdc45 Suggests a Conformational Switch that May Regulate DNA Replication

Read this article at
SUMMARY
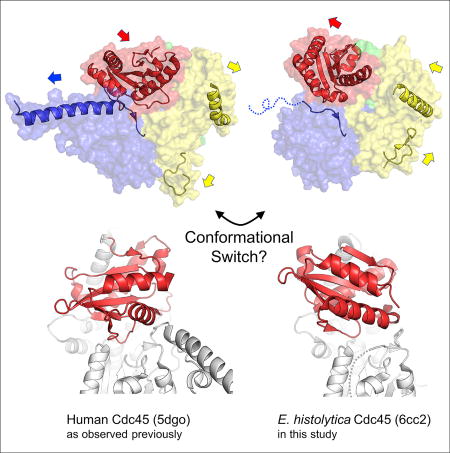
Cdc45 plays a critical role at the core of the eukaryotic DNA replisome, serving as an essential scaffolding component of the replicative helicase holoenzyme Cdc45-MCM-GINS (CMG) complex. A 1.66-Å-resolution crystal structure of the full-length Cdc45 protein from Entamoeba histolytica shows a protein fold similar to that observed previously for human Cdc45 in its active conformation, featuring the overall disk-like monomer shape and intimate contacts between the N- and C-terminal DHH domains. However, the E. histolytica Cdc45 structure shows several unique features, including a distinct orientation of the C-terminal DHHA1 domain, concomitant disordering of the adjacent protruding α-helical segment implicated in DNA polymerase ε interactions, and a unique conformation of the GINS/Mcm5-binding loop. These structural observations collectively suggest the possibility that Cdc45 can sample multiple conformations corresponding to different functional states. We propose that such conformational switch of Cdc45 may allow regulation of protein-protein interactions important in DNA replication.
Graphical Abstract
Related collections
Most cited references20
- Record: found
- Abstract: found
- Article: not found
The PSIPRED protein structure prediction server.
- Record: found
- Abstract: found
- Article: not found
DNA replication in eukaryotic cells.
- Record: found
- Abstract: found
- Article: not found