- Record: found
- Abstract: found
- Article: found
CRISPR-Pass: Gene Rescue of Nonsense Mutations Using Adenine Base Editors

Read this article at
Abstract
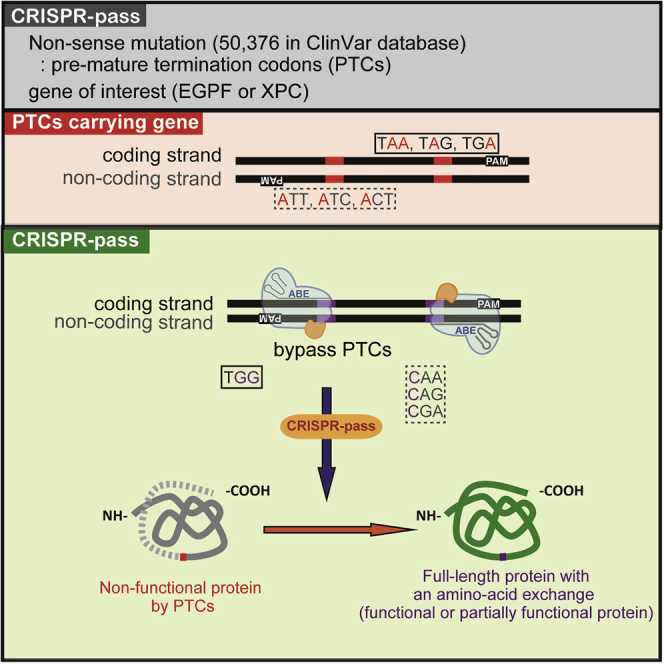
A nonsense mutation is a substitutive mutation in a DNA sequence that causes a premature termination during translation and produces stalled proteins, resulting in dysfunction of a gene. Although it usually induces severe genetic disorders, there are no definite methods for inducing read through of premature termination codons (PTCs). Here, we present a targeted tool for bypassing PTCs, named CRISPR-pass, that uses CRISPR-mediated adenine base editors. CRISPR-pass, which should be applicable to 95.5% of clinically significant nonsense mutations in the ClinVar database, rescues protein synthesis in patient-derived fibroblasts, suggesting potential clinical utility.
Graphical Abstract
Abstract
Lee et al. showed that CRISPR-pass, based on adenine base editors, would be a targeted tool for bypassing premature termination codons. This system could be applicable to ∼95% of clinically significant nonsense mutations, related to genetic diseases, in the ClinVar database.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements

- Record: found
- Abstract: found
- Article: found
Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion
- Record: found
- Abstract: found
- Article: not found