- Record: found
- Abstract: found
- Article: found
Surveillance of Human Astrovirus Infection in Brazil: The First Report of MLB1 Astrovirus

Read this article at
Abstract
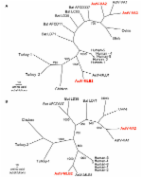
Human astrovirus (HAstV) represents the third most common virus associated with acute diarrhea (AD). This study aimed to estimate the prevalence of HAstV infection in Brazilian children under 5 years of age with AD, investigate the presence of recently described HAstV strains, through extensive laboratory-based surveillance of enteric viral agents in three Brazilian coastal regions between 2005 and 2011. Using reverse transcription-polymerase chain reaction (RT-PCR), the overall HAstV detection rate reached 7.1% (207/2.913) with percentage varying according to the geographic region: 3.9% (36/921) in the northeast, 7.9% in the south (71/903) and 9.2% in the southeast (100/1.089) ( p < 0.001). HAstV were detected in cases of all age groups. Detection rates were slightly higher during the spring. Nucleotide sequence analysis of a 320-bp ORF2 fragment revealed that HAstV-1 was the predominant genotype throughout the seven years of the study. The novel AstV-MLB1 was detected in two children with AD from a subset of 200 samples tested, demonstrating the circulation of this virus both the in northeastern and southeastern regions of Brazil. These results provide additional epidemiological and molecular data on HAstV circulation in three Brazilian coastal regions, highlighting its potential to cause infantile AD.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Multiple novel astrovirus species in human stool.
- Record: found
- Abstract: found
- Article: not found
Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis.
- Record: found
- Abstract: found
- Article: found