- Record: found
- Abstract: found
- Article: found
DNA Binding of the Cell Cycle Transcriptional Regulator GcrA Depends on N6-Adenosine Methylation in Caulobacter crescentus and Other Alphaproteobacteria
Read this article at
Abstract
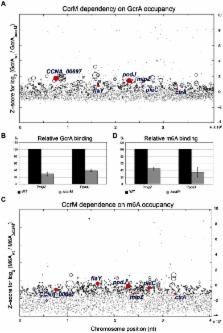
Several regulators are involved in the control of cell cycle progression in the bacterial model system Caulobacter crescentus, which divides asymmetrically into a vegetative G1-phase (swarmer) cell and a replicative S-phase (stalked) cell. Here we report a novel functional interaction between the enigmatic cell cycle regulator GcrA and the N6-adenosine methyltransferase CcrM, both highly conserved proteins among Alphaproteobacteria, that are activated early and at the end of S-phase, respectively. As no direct biochemical and regulatory relationship between GcrA and CcrM were known, we used a combination of ChIP (chromatin-immunoprecipitation), biochemical and biophysical experimentation, and genetics to show that GcrA is a dimeric DNA–binding protein that preferentially targets promoters harbouring CcrM methylation sites. After tracing CcrM-dependent N6-methyl-adenosine promoter marks at a genome-wide scale, we show that these marks recruit GcrA in vitro and in vivo. Moreover, we found that, in the presence of a methylated target, GcrA recruits the RNA polymerase to the promoter, consistent with its role in transcriptional activation. Since methylation-dependent DNA binding is also observed with GcrA orthologs from other Alphaproteobacteria, we conclude that GcrA is the founding member of a new and conserved class of transcriptional regulators that function as molecular effectors of a methylation-dependent (non-heritable) epigenetic switch that regulates gene expression during the cell cycle.
Author Summary
Methylation of genomic DNA at a specific regulatory site can impact a myriad of processes in eukaryotic cells. In bacteria, methylation at the N6 position of adenosine (m6A) is known to mediate a non-adaptive immunity response to protect cells from foreign DNA. While m6A marks are not known to govern expression of cell cycle genes in Gammaproteobacteria, cell cycle transcription in the model alphaproteobacterium Caulobacter crescentus requires the m6A methyltransferase CcrM that introduces m6A marks at GAnTC sequences and the enigmatic factor GcrA. Investigating if a functional and biochemical relationship exists between CcrM and GcrA, we found that CcrM-dependent m6A marks recruit GcrA to the promoters of cell cycle genes in vitro and in vivo and is required for efficient transcription. GcrA interacts with RNA polymerase, explaining how cell cycle transcription is affected. Importantly, m6A-dependent binding is also seen in GcrA orthologs, indicating that this transcriptional regulatory mechanism by CcrM and GcrA is conserved in Alphaproteobacteria.
Related collections
Most cited references24
- Record: found
- Abstract: found
- Article: not found
N6-methyl-adenine: an epigenetic signal for DNA-protein interactions.
- Record: found
- Abstract: found
- Article: not found
MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter.
- Record: found
- Abstract: found
- Article: not found