- Record: found
- Abstract: found
- Article: found
Aggravation of collagen-induced arthritis by orally administered Porphyromonas gingivalis through modulation of the gut microbiota and gut immune system
Read this article at
Abstract
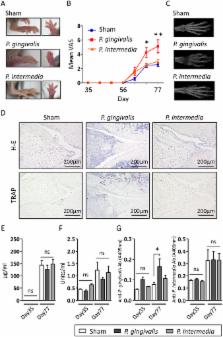
Porhyromonas gingivalis, a causative bacterium of periodontitis, is implicated in the etiology of rheumatoid arthritis (RA), mainly because of expressing peptidyl arginine deiminase (PAD) that generates RA-related autoantigens. However, compared with other periodontopathic bacteria, the precise role of P. gingivalis in RA is largely unknown. We found that orally administered P. gingivalis changed the gut microbiome with concomitant elevation of serum endotoxin and inflammatory markers, and impairment of the gut barrier function. Based on findings showing a relationship between gut microbiota and RA, we investigated whether the change of gut microbiota induced by P. gingivalis and Prevotella intermedia, another periodontopathic bacterium without PAD, is associated with collagen-induced arthritis (CIA). DBA/1J mice were orally administered with or without bacteria followed by induction of CIA. P. gingivalis, but not P. intermedia, administration significantly aggravated arthritis with increased interleukin-17 levels in sera and culture supernatants, increased Th17 cell proportions among mesenteric lymphocytes, and a significant change in the gut microbiome. However, P. gingivalis administration did not elevate the level of anti-citrullinated protein antibody. These results suggest a unique role of P. gingivalis in the link between periodontitis and RA by affecting the gut immune system and the gut microbiota composition.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells.

- Record: found
- Abstract: found
- Article: found
An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis
- Record: found
- Abstract: found
- Article: not found