- Record: found
- Abstract: found
- Article: found
Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms
Read this article at
Abstract
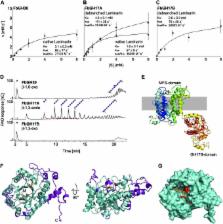
Polysaccharide degradation by heterotrophic microbes is a key process within Earth’s carbon cycle. Here, we use environmental proteomics and metagenomics in combination with cultivation experiments and biochemical characterizations to investigate the molecular details of in situ polysaccharide degradation mechanisms during microalgal blooms. For this, we use laminarin as a model polysaccharide. Laminarin is a ubiquitous marine storage polymer of marine microalgae and is particularly abundant during phytoplankton blooms. In this study, we show that highly specialized bacterial strains of the Bacteroidetes phylum repeatedly reached high abundances during North Sea algal blooms and dominated laminarin turnover. These genomically streamlined bacteria of the genus Formosa have an expanded set of laminarin hydrolases and transporters that belonged to the most abundant proteins in the environmental samples. In vitro experiments with cultured isolates allowed us to determine the functions of in situ expressed key enzymes and to confirm their role in laminarin utilization. It is shown that laminarin consumption of Formosa spp. is paralleled by enhanced uptake of diatom-derived peptides. This study reveals that genome reduction, enzyme fusions, transporters, and enzyme expansion as well as a tight coupling of carbon and nitrogen metabolism provide the tools, which make Formosa spp. so competitive during microalgal blooms.
Related collections
Most cited references25

- Record: found
- Abstract: found
- Article: found
The Proteomics Identifications (PRIDE) database and associated tools: status in 2013
- Record: found
- Abstract: not found
- Article: not found
Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom
- Record: found
- Abstract: found
- Article: not found