- Record: found
- Abstract: found
- Article: found
The Functional Characterization of Long Noncoding RNA SPRY4-IT1 in Human Melanoma Cells
Read this article at
Abstract
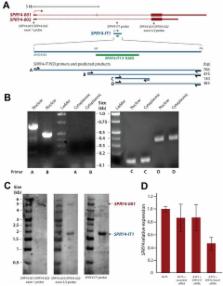
Expression of the long noncoding RNA (lncRNA) SPRY4-IT1 is low in normal human melanocytes but high in melanoma cells. siRNA knockdown of SPRY4-IT1 blocks melanoma cell invasion and proliferation, and increases apoptosis. To investigate its function further, we affinity purified SPRY4-IT1 from melanoma cells and used mass spectrometry to identify the protein lipin 2, an enzyme that converts phosphatidate to diacylglycerol (DAG), as a major binding partner. SPRY4-IT1 knockdown increases the accumulation of lipin2 protein and upregulate the expression of diacylglycerol O-acyltransferase 2 (DGAT2) an enzyme involved in the conversion of DAG to triacylglycerol (TAG). When SPRY4-IT1 knockdown and control melanoma cells were subjected to shotgun lipidomics, an MS-based assay that permits the quantification of changes in the cellular lipid profile, we found that SPRY4-IT1 knockdown induced significant changes in a number of lipid species, including increased acyl carnitine, fatty acyl chains, and triacylglycerol (TAG). Together, these results suggest the possibility that SPRY4-IT1 knockdown may induce apoptosis via lipin 2-mediated alterations in lipid metabolism leading to cellular lipotoxicity.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
The transcriptional landscape of the mammalian genome.
- Record: found
- Abstract: found
- Article: not found
Long noncoding RNA as modular scaffold of histone modification complexes.
- Record: found
- Abstract: found
- Article: not found