- Record: found
- Abstract: found
- Article: found
RNA‐protein interactions in an unstructured context
Read this article at
Abstract
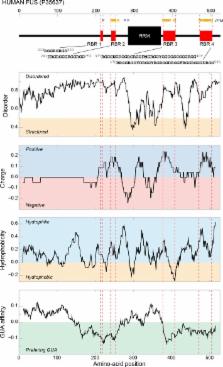
Despite their importance, our understanding of noncovalent RNA–protein interactions is incomplete. This especially concerns the binding between RNA and unstructured protein regions, a widespread class of such interactions. Here, we review the recent experimental and computational work on RNA–protein interactions in an unstructured context with a particular focus on how such interactions may be shaped by the intrinsic interaction affinities between individual nucleobases and protein side chains. Specifically, we articulate the claim that the universal genetic code reflects the binding specificity between nucleobases and protein side chains and that, in turn, the code may be seen as the Rosetta stone for understanding RNA–protein interactions in general.
Related collections
Most cited references71
- Record: found
- Abstract: not found
- Article: not found
The origin of the genetic code.
- Record: found
- Abstract: found
- Article: not found
Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level.

- Record: found
- Abstract: found
- Article: found