- Record: found
- Abstract: found
- Article: found
PLANiTS: a curated sequence reference dataset for plant ITS DNA metabarcoding
Read this article at
Abstract
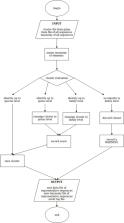
DNA metabarcoding combines DNA barcoding with high-throughput sequencing to identify different taxa within environmental communities. The ITS has already been proposed and widely used as universal barcode marker for plants, but a comprehensive, updated and accurate reference dataset of plant ITS sequences has not been available so far. Here, we constructed reference datasets of Viridiplantae ITS1, ITS2 and entire ITS sequences including both Chlorophyta and Streptophyta. The sequences were retrieved from NCBI, and the ITS region was extracted. The sequences underwent identity check to remove misidentified records and were clustered at 99% identity to reduce redundancy and computational effort. For this step, we developed a script called ‘better clustering for QIIME’ (bc4q) to ensure that the representative sequences are chosen according to the composition of the cluster at a different taxonomic level. The three datasets obtained with the bc4q script are PLANiTS1 (100 224 sequences), PLANiTS2 (96 771 sequences) and PLANiTS (97 550 sequences), and all are pre-formatted for QIIME, being this the most used bioinformatic pipeline for metabarcoding analysis. Being curated and updated reference databases, PLANiTS1, PLANiTS2 and PLANiTS are proposed as a reliable, pivotal first step for a general standardization of plant DNA metabarcoding studies. The bc4q script is presented as a new tool useful in each research dealing with sequences clustering.
Database URL: https://github.com/apallavicini/bc4q; https://github.com/apallavicini/PLANiTS.
Related collections
Most cited references30

- Record: found
- Abstract: found
- Article: found
Validation of the ITS2 Region as a Novel DNA Barcode for Identifying Medicinal Plant Species
- Record: found
- Abstract: not found
- Article: not found
Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data
- Record: found
- Abstract: found
- Article: not found