- Record: found
- Abstract: found
- Article: found
VARIETAL IDENTIFICATION IN HOUSEHOLD SURVEYS: RESULTS FROM THREE HOUSEHOLD-BASED METHODS AGAINST THE BENCHMARK OF DNA FINGERPRINTING IN SOUTHERN ETHIOPIA
Read this article at
Abstract
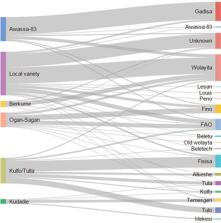
Accurate crop varietal identification is the backbone of any high-quality assessment of outcomes and impacts. Sweetpotato ( Ipomoea batatas) varieties have important nutritional differences, and there is a strong interest to identify nutritionally superior varieties for dissemination. In agricultural household surveys, such information is often collected based on the farmer’s self-report. In this article, we present the results of a data capture experiment on sweet potato varietal identification in southern Ethiopia. Three household-based methods of identifying varietal adoption are tested against the benchmark of DNA fingerprinting: (A) Elicitation from farmers with basic questions for the most widely planted variety; (B) Farmer elicitation on five sweet potato phenotypic attributes by showing a visual-aid protocol; and (C) Enumerator recording observations on five sweet potato phenotypic attributes using a visual-aid protocol and visiting the field. In total, 20% of farmers identified a variety as improved when in fact it was local and 19% identified a variety as local when it was in fact improved. The variety names given by farmers delivered inconsistent and inaccurate varietal identities. Visual-aid protocols employed in methods B and C were better than those in method A, but greatly underestimated the adoption estimates given by the DNA fingerprinting method. Our results suggest that estimating the adoption of improved varieties with methods based on farmer self-reports is questionable and point towards a wider use of DNA fingerprinting in adoption and impact assessments.
Related collections
Most cited references27

- Record: found
- Abstract: found
- Article: found
A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species
- Record: found
- Abstract: found
- Article: not found
Green revolution: impacts, limits, and the path ahead.
- Record: found
- Abstract: found
- Article: not found