- Record: found
- Abstract: found
- Article: not found
A Conformational Investigation of Propeptide Binding to the Integral Membrane Protein γ-Glutamyl Carboxylase Using Nanodisc Hydrogen Exchange Mass Spectrometry

Read this article at
Abstract
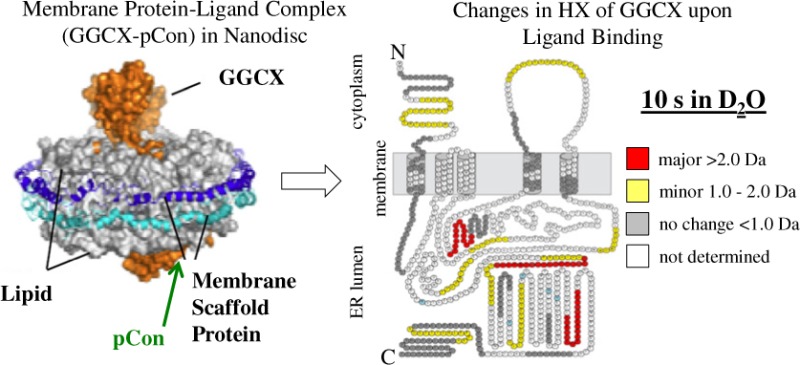
Gamma (γ)-glutamyl carboxylase (GGCX) is an integral membrane protein responsible for the post-translational catalytic conversion of select glutamic acid (Glu) residues to γ-carboxy glutamic acid (Gla) in vitamin K-dependent (VKD) proteins. Understanding the mechanism of carboxylation and the role of GGCX in the vitamin K cycle is of biological interest in the development of therapeutics for blood coagulation disorders. Historically, biophysical investigations and structural characterizations of GGCX have been limited due to complexities involving the availability of an appropriate model membrane system. In previous work, a hydrogen exchange mass spectrometry (HX MS) platform was developed to study the structural configuration of GGCX in a near-native nanodisc phospholipid environment. Here we have applied the nanodisc–HX MS approach to characterize specific domains of GGCX that exhibit structural rearrangements upon binding the high-affinity consensus propeptide (pCon; AVFLSREQANQVLQRRRR). pCon binding was shown to be specific for monomeric GGCX-nanodiscs and promoted enhanced structural stability to the nanodisc-integrated complex while maintaining catalytic activity in the presence of carboxylation co-substrates. Noteworthy modifications in HX of GGCX were prominently observed in GGCX peptides 491–507 and 395–401 upon pCon association, consistent with regions previously identified as sites for propeptide and glutamate binding. Several additional protein regions exhibited minor gains in solvent protection upon propeptide incorporation, providing evidence for a structural reorientation of the GGCX complex in association with VKD carboxylation. The results herein demonstrate that nanodisc–HX MS can be utilized to study molecular interactions of membrane-bound enzymes in the absence of a complete three-dimensional structure and to map dynamic rearrangements induced upon ligand binding.
Related collections
Most cited references56
- Record: found
- Abstract: found
- Article: not found
Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation.
- Record: found
- Abstract: found
- Article: not found
High-speed and high-resolution UPLC separation at zero degrees Celsius.
- Record: found
- Abstract: found
- Article: not found
