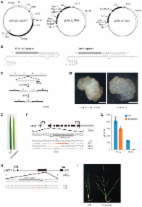
Dear Editor, Genome editing of model organisms is essential for gene function analysis and is thus critical for human health and agricultural production. The current technologies used for genome editing include ZFN (zinc-finger nuclease), meganucleases, TALEN (Transcription activator-like effector nucleases), etc. 1 . These technologies can generate double stranded breaks (DSBs) to either disrupt gene function through generation of premature stop codons by non-homologous end joining (NHEJ) pathway, or to facilitate gene targeting through homologous recombination (HR) with an incoming template. Recently, a new technology for genome editing, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas (CRISPR-associated) systems, has been developed 2 . CRISPR/Cas systems are adaptive defense systems in prokaryotic organisms to fight against alien nucleic acids 3 . The spacer sequences acquired from foreign DNA are positioned between host repeats, and transcribed together as CRISPR RNA (crRNA). In the type II CRISPR system, a single nuclease Cas9, guided by a dual-crRNA:tracrRNA, is sufficient to cleave cognate DNA homologous to the spacer 2 . Efficient cleavage also requires the presence of protospacer adjacent motif (PAM) 5′-NGG-3′ following the spacer sequence. The dual-crRNA:tracrRNA has been further streamlined to a single RNA chimera, called sgRNA (single guide RNA) 2 . Compared with protein-guided technologies, CRISPR/Cas system is much easier to implement, as only short guide RNAs need to be customized to target the genes of interest. Up to now, the CRISPR/Cas system has been successfully applied to efficient genome editing in many eukaryotic organisms including human 1 , mice 4 , zebra fish 5 , fly 6 , worm 7 , and yeast 8 . However, the application of CRISPR/Cas system in plants has not been reported. Rice (Oryza sativa L.) is a major staple crop in the grass family (Poaceae), feeding half of the world's population. Rice is also used as a model monocot plant for biological studies because it has a relatively small genome compared to other cereal crops and is easy to be manipulated genetically. We demonstrate in this study that the CRISPR/Cas technology can achieve efficient targeted mutagenesis in transgenic rice. Our work paves the way for large-scale genome editing in rice, which is important for quality improvement and yield increase of rice. To accommodate the CRISPR/Cas system to Agrobacterium-mediated plant transformation, we designed Gateway™ binary T-DNA vectors for co-expression of CAS9 and guide RNA (either sgRNA or dual-crRNA:tracrRNA, see Figure 1A). Gene-specific spacer sequence was cloned into entry vectors for expression of guide RNA (Supplementary information, Figure S1 and Data S1), which was then cloned into destination vectors containing the CAS9 expression cassette. To reconstitute Cas9 ribonucleoprotein complex in the nucleus, Cas9 was attached with a nuclear localization signal, and guide RNAs were driven by pol III type promoter of U3 snRNA. CAS9 coding sequence was codon-optimized for expression in rice (Supplementary information, Figure S2), and was driven by the maize Ubiquitin (Ubi) promoter. To facilitate Cas9 binding and R-loop formation, we chose the sgRNA with the secondary structure containing a dangling spacer, an extended hairpin region and a long 3′ end (Figure 1B). To test whether the CRISPR/Cas system can be applied in plant cells, we first investigated whether the system can generate DSB in rice callus using the DGU.US as a reporter (Supplementary information, Figure S3). GUS activity can be restored through single strand annealing (SSA) upon DNA cleavage between the repeat regions in the DGU.US reporter 9 (Figure 1C). We designed both sgRNA and dual-crRNA:tracrRNA to target the reporter. The result showed that strong GUS staining spots were detected in rice calli after the CRISPR/Cas system and the reporter were co-transformed through particle bombardment, whereas no GUS signal was observed in those transformed with the reporter alone (Figure 1D). Both sgRNA and dual-crRNA:tracrRNA were effective in this assay, indicating that pre-crRNA can be properly processed in rice (plant) cells. To further test whether the CRISPR/Cas technology can be used to specifically disrupt an endogenous gene in rice, we designed sgRNAs and dual-crRNA:tracrRNAs targeting either the second exon of the CHLOROPHYLL A OXYGENASE 1 (CAO1) gene or the third exon of the LAZY1 gene, and transformed them into Kitaake, a japonica rice variety with short life cycle 10 . The seedlings of the loss-of-function mutant cao1 show a pale green leaf phenotype due to defective synthesis of Chlorophyll b (Chl b), which is easily observed at an early developmental stage, whereas the la1 mutant, loss-of-function mutant of LAZY1 gene, exhibited a pronounced tiller-spreading phenotype that can be observed after tillering stage 11 . We obtained 30 independent transgenic lines for sgRNA construct and 45 lines for dual-crRNA:tracrRNA construct for the CAO1 gene. We found that some transgenic plants displayed pale green leaf blades (Figure 1E), and we conducted genotyping analysis on these lines using gene-specific primers. The result showed that either loss of peak/gain of peak or overlapping peak around the target site was observed in the sequencing chromatograms (Figure 1F and Supplementary information, Figure S4), which confirmed mutations in the CAO1 gene. The occurrence of indels at 3-4-bp upstream of PAM is consistent with the location of Cas9 cleavage site. We randomly selected transgenic lines with biallelic mutation, and measured the contents of Chl a and Chl b 12 . The result showed that Chl b content was reduced to a marginal level, consistent with the cao1 phenotype (Figure 1G). We obtained only 12 independent transgenic lines for sgRNA construct for LAZY1 gene. Sequencing analysis showed that 11 out of 12 lines bear mutations in the specific region of LAZY1 gene, confirming the disruption of LAZY1 gene (Figure 1H and Supplementary information, Figure S5). Appearance of tiller-spreading phenotypes in 6 homogenous mutation lines further supported the conclusion (Figure 1I). Notably, in the case of sgRNA, about 83.3% and 91.6% of the independent lines of the T1 transgenic rice beared mutations in CAO1 and LAZY1, respectively, among which 4 lines (13.3%) for CAO1 and 6 lines (50%) for LAZY1 beared biallelic mutations (Supplementary information, Table S1). These results indicate that transgenic rice with mutated gene of interest can be easily generated in the T1 generation by using CRISPR/Cas technology, and that the high efficiency of targeted mutagenesis can be achieved in different genes. This high efficiency is possibly due to the unique feature of the CRISPR/Cas system, i.e., different from ZFN or TALEN, Cas9-mediated cleavage tolerates DNA methylation 13 . This gives the CRISPR/Cas technology leverage for genome editing in plants with high GC content in the genome such as rice. It seems that sgRNA constructs generate targeted mutagenesis more efficiently than dual-crRNA:tracrRNA constructs (Supplementary information, Table S1), the mechanism of which needs to be investigated in the future. To facilitate selection of spacers to target other rice genes, we also used criteria reported recently, which exclude potential mismatches of the first 8 nucleotides of the 20-nt spacer region and leaky activity on 5′-NAG-3′ PAM 8 , and designed more than 3.6 million spacers (Supplementary information, Data S2). We showed that transgenic rice with mutations in specific genes could be generated through the CRISPR/Cas technology in a straightforward manner. Our strategy for vector construction is modular, efficient, and expandable for multiplex gene editing. Because dual expression of pre-crRNA:tracrRNA can be correctly processed in the plant cells although the mutagenesis efficiency is relatively low in the two genes that we tested (Figure 1 and Supplementary information, Table S1), the entry vectors that we designed has the potential to host two sgRNAs or multiple spacers, in an array of crRNA. The facile genome editing at specific sites in rice will speed up functional characterization of rice genes, especially for those genes with family members, and will greatly promote the effort to improve rice quality and yield through agricultural biotechnology. The compiled list of the specific spacers for each gene in rice genome will also help to accelerate the application of the CRISPR/Cas system in rice. Further work will be focused on whether the CRISPR/Cas system can be applied successfully to other cereals with bigger and more complicated genomes such as maize and wheat. Meanwhile, further modification of the CRISPR/Cas system, e.g., removal of the components of the CRISPR/Cas system after the target genes are mutated, will promote the application of this new technology in agriculture.